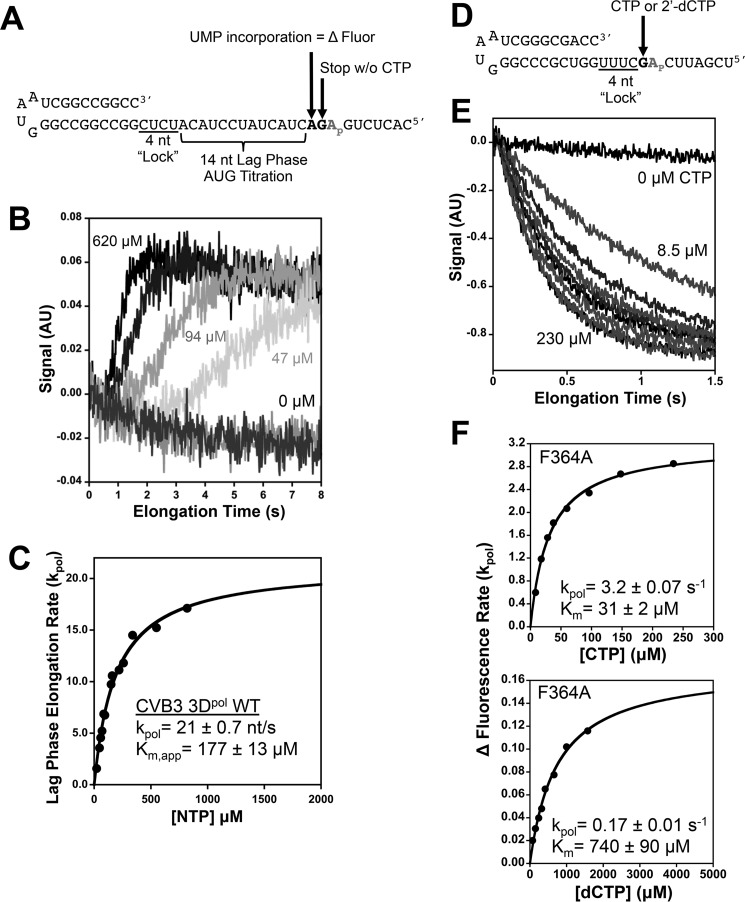
FIGURE 3.
Processive elongation and single nucleotide incorporation measured by stopped-flow 2-aminopurine fluorescence. A, hairpin primer-template RNA on which preinitiated elongation complexes are assembled by only supplying GTP and ATP in the reaction, causing them to stall at a +4 product. Stopped-flow addition of ATP + GTP + UTP then results in rapid elongation to the +18 product where the complex stalls because CTP is not present. This translocates a unique 2-aminopurine base analog (Ap) into the +2 binding pocket on the polymerase where its fluorescence increases because it is fully unstacked from both neighboring bases. B, stopped-flow traces showing the shortening of the lag phase that reflects faster processive elongation as the NTP concentration is increased. C, curve fitting of the rates extracted from the lag phase versus NTP concentration allows the determination of processive elongation rates and Km values. D, structure of the hairpin primer-template RNA used for single nucleotide incorporation assays where preinitiation with ATP and GTP result in stalled elongation complexes with the template strand 2-aminopurine in the +2 position. E, stopped-flow traces demonstrating the single step quenching of fluorescence as 2-aminopurine is translocated from the +2 position to the +1 position. F, analysis of CVB3 3Dpol F364A single nucleotide turnover rates as a function of CTP (top) and 2′-dCTP (bottom) concentrations. AU, arbitrary units; nt, nucleotide(s).