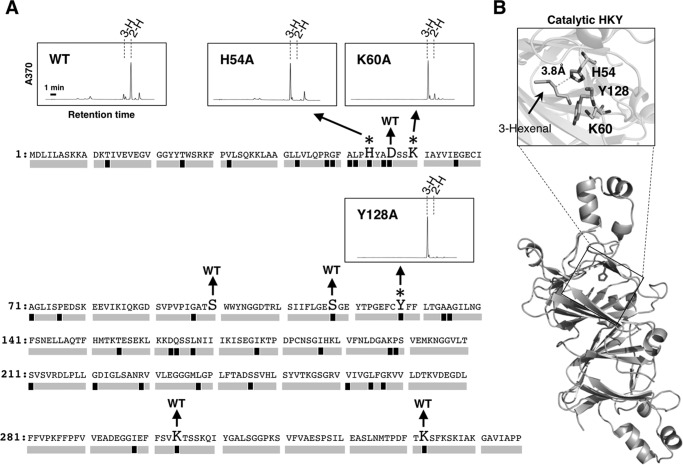
FIGURE 4.
Determination of catalytic amino acids of HI. A, effect of point mutation on HI activity. Amino acids written in large print indicate point-muted amino acids, and chromatograms of the point-muted proteins completely losing activity are shown. Asterisks suggest essential amino acids to show the HI activity, and point-mutated amino acids with no effect on the HI activity are indicated by WT. Black boxes under the amino acid sequence of CaHI indicate amino acids conserved in all proteins belonging to Solanaceae HI clade but not in HI-like clade. Gray boxes show amino acids not conserved in Solanaceae HI clade or conserved both HI and HI-like clades. 3-H and 2-H in chromatograms indicate peaks of (Z)-3- and (E)-2-hexenal-DNPs, respectively. B, homology modeling of CaHI to deduce catalytic amino acids. Catalytic amino acids locate in the same pocket and near substrate (Z)-3-hexenal. PDB ID of template protein, identity between CaHI and template protein, and QMEAN score are 2e9q, 23%, and 0.62, respectively.