FIGURE 8.
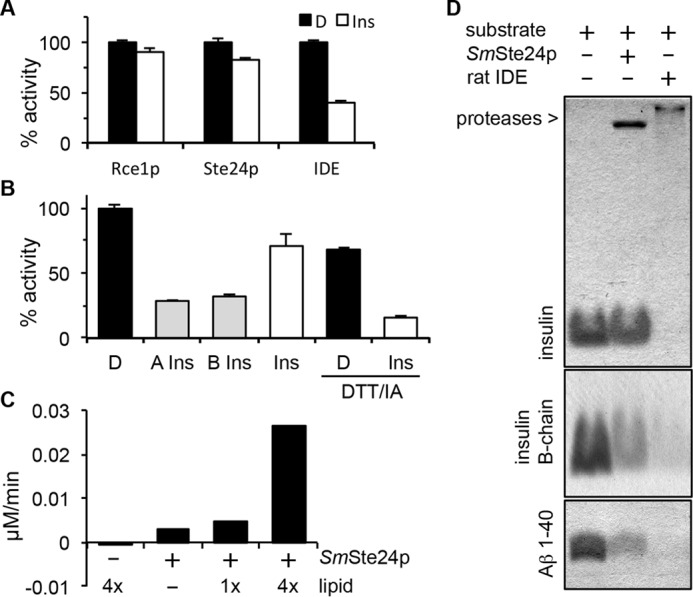
Evaluation of Ste24p activity against biological peptides. A, competition assays were performed using the indicated protease and appropriate FRET substrate in the absence or presence of insulin. See Fig. 3A for Rce1p and Ste24p substrates and Fig. 4A for the IDE substrate. Substrates were used at 10 μm, and competing peptide was used at 40 μm. D, DMSO; Ins, whole insulin. B, competition assays were performed as described for A using Ste24p membranes and the indicated insulin species. DTT/IA, pretreatment of the competitor with a 3-fold molar excess of DTT, followed by a 6-fold molar excess of iodoacetamide (IA). This serves to dissociate insulin A- and B-chains and block reformation of disulfide bonds. D, DMSO; A Ins, carboxymethylated insulin A-chain; B Ins, carboxymethylated insulin B-chain; Ins, whole insulin. The graphs for A and B are representative of multiple independent experiments (three in each case). The specific graphs represent the averaged value of replicates (n = 2) from a single experiment where the DMSO-treated control was set to 100% activity; error bars represent the range. C, purified SmSte24p (0.8 μm) was incubated with the Aβ 1–28 FRET reporter (50 μm) in the absence and presence of E. coli lipid. 1x, 125 μg/ml total E. coli lipid. rfu, relative fluorescence units. The graph is representative of five independent tests of lipid effects on this enzyme/substrate combination. D, SDS-PAGE analysis of products derived from incubation of purified SmSte24p or rat IDE with indicated substrates in the presence of E. coli lipid. The gels are representative of five independent experiments. The concentrations of enzymes, substrates, and lipids used are described under “Experimental Procedures.”
