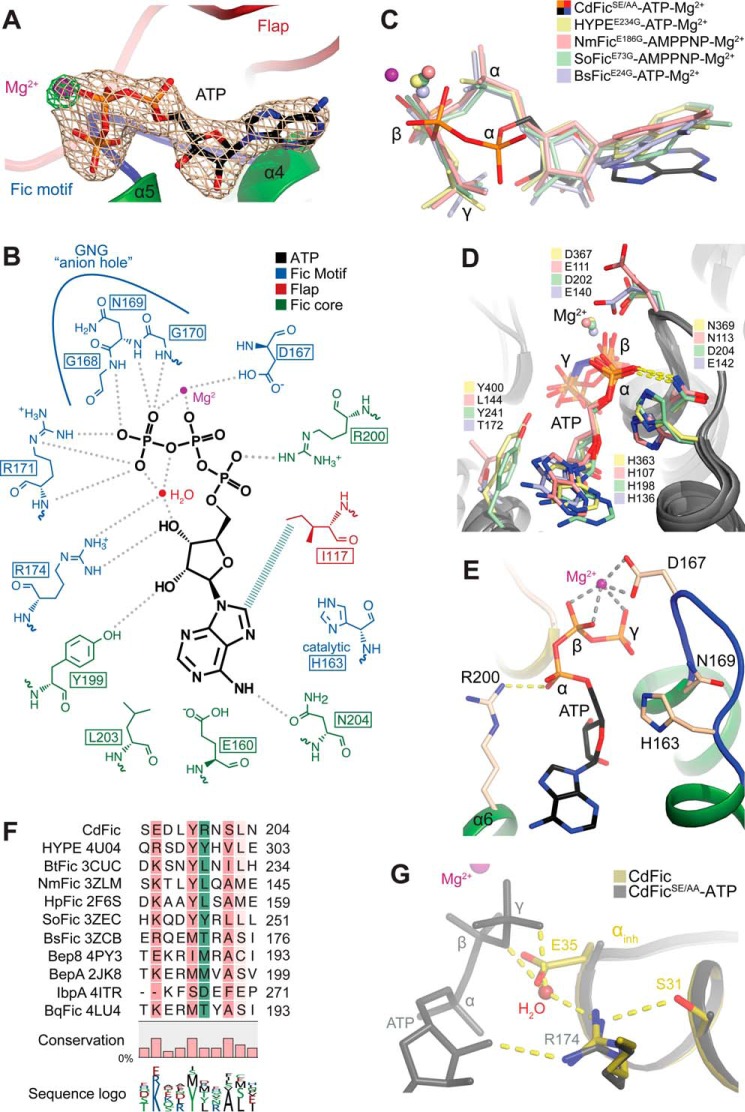
FIGURE 2.
The ATP binding site. A, the CdFicSE/AA-ATP binding pocket. An unbiased Fo − Fc electron density omit map around ATP is contoured at 4 σ and shown in gray. An Fo − Fc electron density omit map around Mg2+ ion is shown in green and contoured at 6.5 σ to emphasize the strong Mg2+ peak. Mg2+ is shown in purple, and the Fic motif and the flap are shown in blue and red, respectively. B, schematic diagram of the CdFicSE/AA-ATP coordination as determined by LIGPLOT (63). C, superposition of ATP from CdFicSE/AA-ATP crystal structure and the structures of HYPEE234G-ATP (H. sapiens; Protein Data Bank code 4U07), NmFicE186G-AMPPNP (N. meningitides; Protein Data Bank code 3ZLM), SoFicE73G-AMPPNP (Shewanella oneidensis; Protein Data Bank code 3ZEC), and BsFic (VbhT-VbhAE24G-ATP) (B. schoenbuchensis; Protein Data Bank code 3ZCB). D, the ATP binding mode of Fic domain structures from C not including CdFicSE/AA. E, the ATP binding mode of CdFicSE/AA highlighting the salt bridge between Arg-200 and α-phosphate shown in identical orientation as D. F, structurally based multiple sequence alignment of Fic domains. Fic domains with the indicated Protein Data Bank accession codes were aligned using the program Strap (64), and the alignment was calculated based on the structural superposition and visualized using the CLC Main Workbench 7.5.1. (Qiagen Bioinformatics). The conservation below 30% identity is not colored, but a progressively darker red color represents conservation in the 30–100% range. The position of Arg-200 residue is shown with green boxes. The fractional conservation is represented below the alignment using bars and sequence conservation with sequence logo. G, superposition of the inhibitory motif residues Ser-31/Gluinh-35 in CdFic (dark yellow) with the CdFicSE/AA-ATP structure (gray). The ATP from the CdFicSE/AA-ATP structure is colored in gray, and a water molecule from the CdFicSE/AA-ATP structure is shown in red color interacting with both the γ-phosphate and the Fic motif Arg-174 and is in an overlapping position with the Gluinh-35 side chain of the CdFic structure.