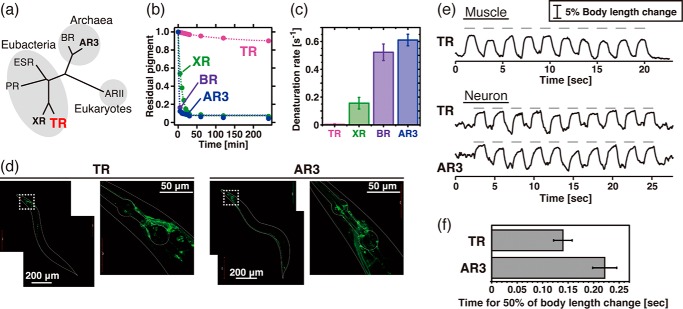
FIGURE 1.
Thermal stability and optogenetic availability of TR. a, phylogenetic tree of microbial proton-pumping rhodopsins of known structure. PR, ESR, and ARII stand for proteorhodopsin, Exiguobacterium sibiricum rhodopsin, and Acetabularia rhodopsin II, respectively. b, denaturation kinetics of TR, XR, BR, and AR3 at 75 °C. The broken lines represent fitting curves of a single exponential function. c, denaturation rates at 75 °C estimated from the kinetics shown in b (n = 3). d, expression of TR::GFP and AR3::GFP in transgenic C. elegans by the pan-neuronal promoter. e, paralyzing activity of TR and AR3. Upper panel, elongation of the body length of C. elegans worms expressing TR::GFP in body wall muscle cells (n = 7). Ten pulses of green light (0.8 milliwatt/mm2, 550 nm) of 1.0 s in duration with 1.0-s interstimulus intervals were applied as indicated by the gray bars. Lower panel, elongation of the body length of worms expressing either TR::GFP (n = 4) or AR3::GFP (n = 4) in neurons with 10 green pulses (4.4 milliwatts/mm2, 550 nm) of 1.0 s with 1.0-s interstimulus intervals. f, the average time for the half of the body length change in worms expressing TR and AR3 calculated from the data shown in e, lower panel.