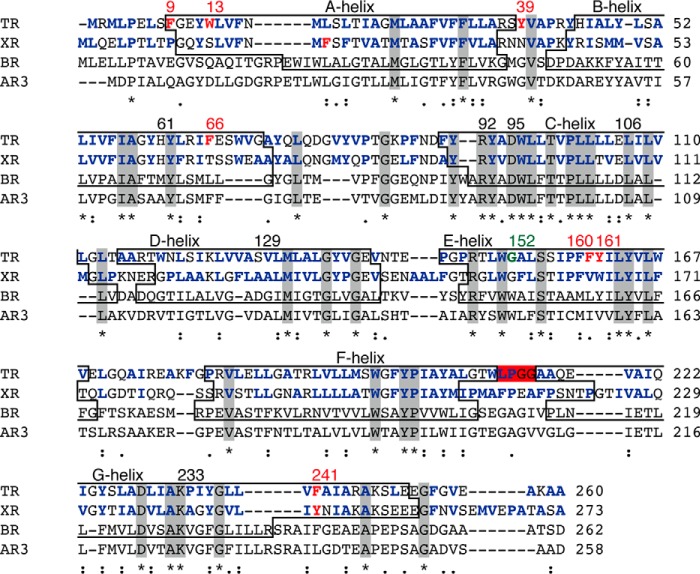
FIGURE 2.
Amino acid sequence alignments of microbial proton-pumping rhodopsins generated by MUSCLE (multiple sequence comparison by log-expectation). Identical residues among these molecules are shown in gray boxes and by asterisks. The transmembrane helices (A-to-G helices), are shown in boxes. The helices belonging to AR3 are not shown because its crystal structure is lacking. The hydrophobic residues of TR and XR are shown in bold blue letters. TR and XR have non-identical aromatic residues, shown in bold red letters, and involve the aromatic-aromatic interactions. The LPGG sequence in the FG loop is shown in a red box. The conserved residue Gly152, which is assumed to contribute to secondary chromophore binding in TR, is highlighted in bold green letters. The residual numbers of TR are shown at the top of the sequence.