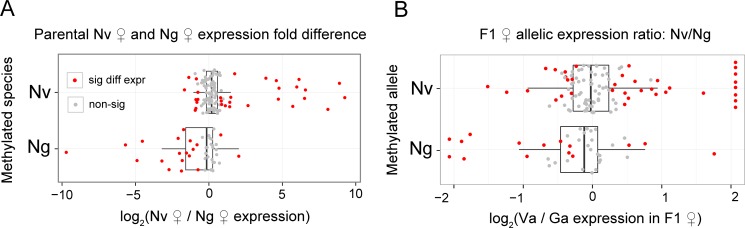
Fig 5. Differentially methylated genes between Nasonia vitripennis (Nv) and Nasonia giraulti (Ng) display a bias toward methylated alleles for both total and allelic expression levels.
(A) Boxplot of parental Nv/Ng expression ratios for differentially methylated genes. Criteria are provided in materials and methods. Differentially expressed genes (FDR < 0.05) are shown in red. Among these genes, the methylated species show significantly higher expression compared to the species with the non-methylated allele. (B) Boxplot of F1 allele-specific expression ratios for differentially methylated genes between Nv and Ng, quantified by Nv allelic expression divided by Ng allelic expression in F1s (x-axis). Criteria are provided in materials and methods. Genes that are monoallelically expressed in Nv (Ng) were plotted at the right (left), because their log2 value is undefined. Genes with significant allelic expression bias (FDR < 0.01) are shown in red, and their methylation patterns are significantly biased toward the highly expressed alleles. Data presented in this figure can be found at http://dx.doi.org/10.5061/dryad.qf2t8.