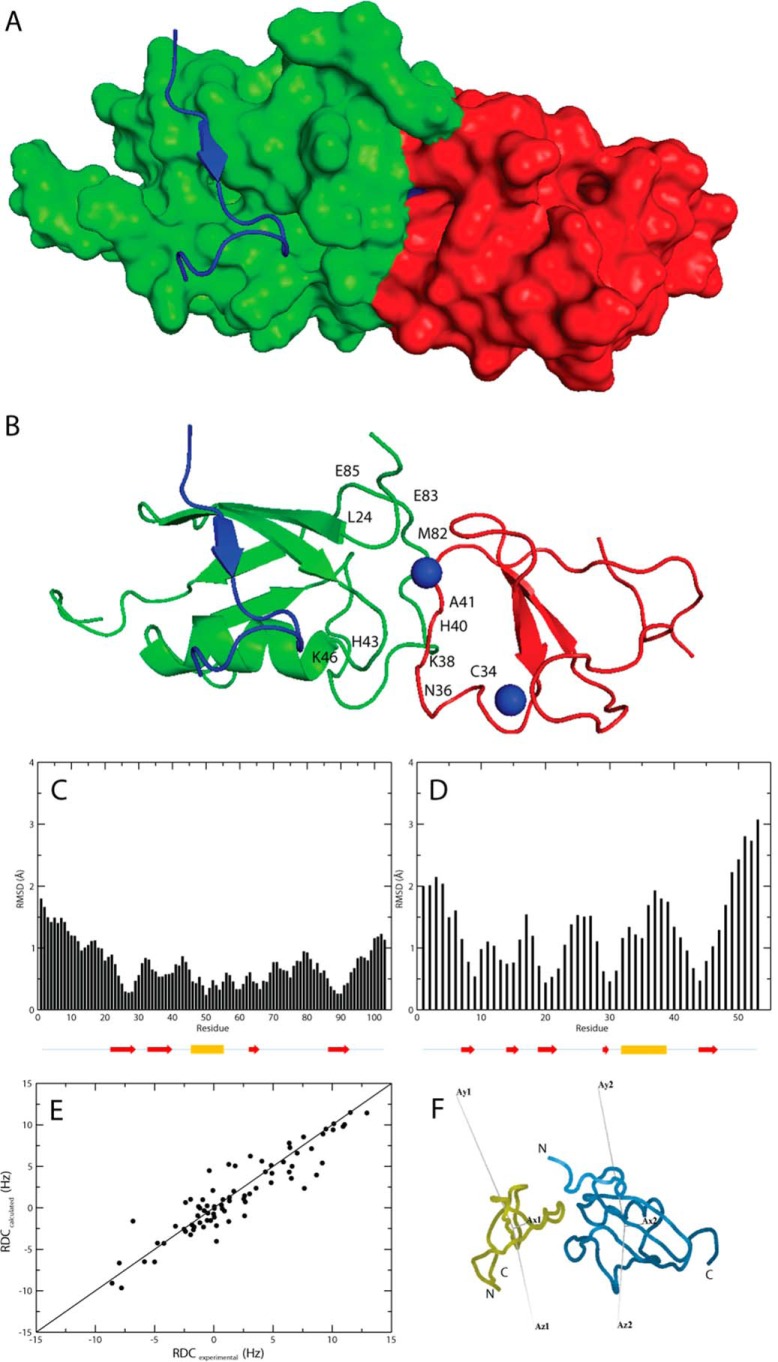
FIGURE 3.
A, model of the ZZ domain-SUMO1 complex shown in a surface representation made using PyMOL (44). SUMO1 is colored green, the ZZ domain is colored red, and the peptide corresponding to a SIM motif is colored in blue (Protein Data Bank code 2ASQ; Ref. 6). B, model of the ZZ domain-SUMO1 complex shown in a ribbon representation using the same color scheme as in A with the two zinc ions depicted as blue spheres. The residues in SUMO1 and the ZZ domain affected by the interaction in the NMR epitope mapping experiments are indicated in the model, corresponding to the same residues shown for the individual protein models in Fig. 1, C and D. C, per residue r.m.s.d. between interface residues and backbone Cα atoms in the modeled complex for SUMO1 residues 1–103. The location for the secondary structure elements of SUMO1 are indicated by arrows (red) for β-strands and a cylinder for the single α-helix (yellow). D, per residue r.m.s.d. between interface residues and backbone Cα atoms in the modeled complex for the ZZ domain residues 1–53. The location for the secondary structure elements of the ZZ domain is indicated by arrows (red) for β-strands and a cylinder for the helical segment (yellow). E, experimental RDCs plotted versus calculated RDCs for the modeled protein complex. F, orientations of the RDC alignment tensor for the ZZ domain and SUMO1 in the ZZ domain-SUMO1 complex in which tensor orientations were fitted using Module (45). The ZZ domain (yellow) and SUMO1 (blue) are shown in ribbon representations.