Preface
In order to engage in adaptive symbioses or genetic exchange, organisms must interact with foreign, non-self elements despite the risks of predation and parasitism. By surveying the interface between self and non-self, immune systems can help ensure the benevolence of these interactions without isolating their hosts altogether. In this Essay, we examine prokaryotic restriction-modification and CRISPR–Cas activities and discuss their analogy to mammalian immune pathways. We further explain how their capacities for resistance and tolerance are optimized to reduce parasitism and immunopathology during encounters with non-self.
Introduction
Across all domains of life, the fitness of an organism may be threatened by parasitic interactions with foreign elements. Accordingly, organisms have evolved a variety of biological barriers which aid in the defense against parasitism. Physical barriers, such as the cellular envelope and those at the surface of mammalian skin and mucosal linings, repel foreign elements rather indiscriminately1, 2. Immune systems, on the other hand, offer the potential to repel particular target elements whilst tolerating others, and may thus be said to function as highly selective barriers. However, the balance of selectivity must be well-calibrated when relying on active means of surveillance and resistance; a system that is too selective risks evasion by pathogens, while one that is unselective risks damaging the host organism through immunopathology. In this regard, the mammalian immune system serves as an excellent example, with innate and adaptive activities3 that are optimized to reliably combat parasitism and generally tolerate the host’s constituents.
It has been appreciated that two prokaryotic systems, known as Restriction–Modification (R–M) and Clustered, Regularly Interspaced, Short Palindromic Repeats in conjunction with Cas proteins (CRISPR–Cas), resemble the mammalian immune system in their ability to actively resist infectious elements with a formidable degree of selectivity, as well as in their capacity for tolerance4–6. Both systems achieve selectivity via nucleic acid surveillance within the cell, where they can assist prokaryotes in sorting through genetic material encountered through horizontal gene transfer (HGT). Although mammalian lineages largely forgo HGT in favor of sexual reproduction, they retain the ability to harbour foreign genetic information through their symbiotic interactions with the microbiota7. Collectively, the genetic repertoire of a multicellular organism, along with the microbiomes of its resident microbiota, can be conceptualized as a metagenome (Box 1). In this Essay, we adopt a broad view of immune systems as selective barriers to parasitism which moderate, but do not prevent, the natural flux of their host’s genomic or metagenomic content. We begin by discussing the principal genetic factors that allow for genome evolution in each domain of life, in order to establish our rationale for the comparison between prokaryotic and mammalian immune systems that follows. By exploring a viewpoint from which we believe the analogy of these systems is most evident, we hope to strengthen general conceptualizations regarding the implications of immune selectivity and emphasize the contextual value of tolerating foreign elements.
Box 1. Microbiomes: pieces of a metagenomic puzzle?
Although an organism may be defined by its genomic content in a strict genetic sense, the classical evolutionary definition is concerned most with its phenotype21. The descriptive power of a genome sequence is therefore limited, in part, to the extent that phenotypes can vary independently of a particular genotype. However, phenotypic variability can conceivably be accounted for, given a better understanding of how multicellular organisms are shaped by environmental factors — including the microbiota that colonize them120–124. Hence, there is increasing effort to define metagenomic information, such as microbiomes, that could be correlated with organismal phenotypes125. Microbiome sequencing efforts attempt to catalogue the genetic repertoires of entire microbial communities126. Analyses of host-associated microbiomes have revealed substantial species- and strain-level diversity, as well as a striking degree of genic diversity. Notably, one major study found that gut microbiomes derived from 138 human stool samples contain a cumulative total of over 5.1 million non-redundant genes — more than 200 times the number of known human genes127. A similar dataset comprising gut microbiomes from 124 individuals had a cumulative total estimated at 3.3 million non-redundant genes, with each microbiome sampling over half a million of these genes and at least 160 different species of bacteria128. Importantly, significant variation has been observed both between individuals and in the same individuals sampled over time129, 130. Moreover, specific microbiome compositions have now been associated with different states of health or disease131–134 — in line with the notion that microbiomes may be viewed as extensions of a multicellular organism’s genomic information135, 136, and perhaps not surprising, given that microbiota constitute an estimated 90% of the cells found in a human body137. It should be noted, however, that none of these human metagenomic analyses to date have considered the host’s sequences. Furthermore, because the contents of a microbiome are not fixed with respect to the host’s nuclear genome, a thorough understanding of human metagenomes will likely demand knowledge of how ecological factors126, 138, in addition to host genotypes139, influence microbiome composition and stability.
Vectors of genome evolution
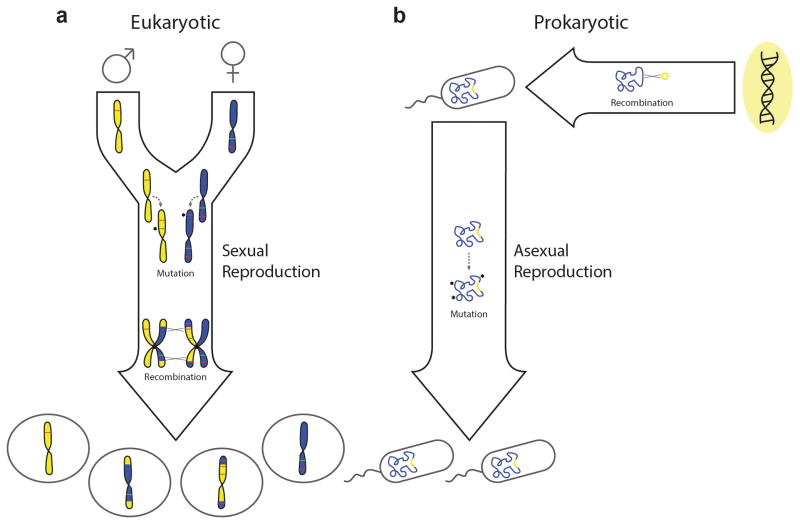
The ‘vertical’ transfer of a genome from parent to progeny during reproduction can result in stochastic alteration of its sequence content via mutation. Mutations are a source of genetic variation and may give rise to novel adaptive genotypes that can be selected for during evolution. However, mutations occurring at random are more likely to be deleterious than beneficial; and, according to Muller’s ratchet mechanism8, 9, genomic degeneration via random mutation can drive an organism to extinction in the absence of a recombination process to increase the chance of reconstructing fully functional genomes10. This fate may be averted through sexual recombination, as it is exhibited in extant eukaryotes (FIG. 1a). Moreover, sexual recombination offers an additional mechanism for generating novel genotypes during reproduction, but it has evolved with reproductive barriers which help to moderate the variation produced by ensuring that genetic exchanges occur between closely related genomes; for example, within a species11. In contrast, recombination can occur between more distantly related genomes through ‘horizontal’ gene transfer (HGT), which constitutes another key avenue for genomic diversification12, 13.
Figure 1. Genome evolution resulting from vertical or horizontal transmission of DNA.
a | Genome evolution in a unicellular and haploid eukaryote that reproduces sexually. Mutation can generate novel alleles (brown lines) within chromosomes during DNA replication, which may be either deleterious or beneficial. Recombination between homologous chromosomes can occur during a tetraploid zygotic stage, prior to any cellular division. Sexual recombination in this manner provides opportunities for the removal of deleterious alleles (red lines), as well as for the introduction of beneficial alleles (green lines). Both processes occur vertically during reproduction. b | Genome evolution in a haploid and unicellular prokaryote that reproduces asexually. Mutation may once again generate novel alleles vertically during reproduction; however, recombination with exogenous sources of DNA must occur through horizontal processes which are not tied to reproduction.
Broadly speaking, HGT refers to the transmission of genetic material from one organism to another, through processes that are not tied to reproduction. Following physical transport into the recipient organism, donor DNA may be incorporated into the genome via recombination with the chromosome(s), or via autonomous replication as an episome. Evidence for HGT in eukaryotes has accumulated over the years, but most reports involve free-living protists and fungi, or endosymbionts inhabiting the cytosol of multicellular organisms13, 14. Meanwhile, the nuclear genome of mammals appears to be well-insulated from HGT, and this has been suggested to result from isolation of their germline cells in the gonads15. One could further speculate that this represents an evolutionary strategy which helps to preserve genomic fitness in mammalian lineages, given that HGT can introduce deleterious mutations in addition to those generated vertically and might be superfluous if sufficient recombination and genetic variation is already achieved through sexual reproduction. However, for unicellular, asexually reproducing organisms such as prokaryotes, HGT provides an avenue for recombination which can in theory operate like sex in averting Muller’s ratchet16, in addition to providing genomic variation (FIG. 1b). Sequencing of diverse prokaryotic phyla and comparative genome analyses suggest the virtual ubiquity of HGT, as well as its potential to drive rapid genome evolution via the acquisition of novel genes en bloc17, 18. Notably, HGT appears to be a common route through which evolving pathogens acquire particular traits, including resistance to antibiotics19. Nevertheless, in the absence of reproductive barriers, the risk of introducing deleterious information into the genome is theoretically greater with HGT than with conventional sex. For example, HGT can introduce mobile genetic elements (MGEs) from unrelated organisms—such as transposons, viruses, or plasmids—which can in turn parasitize the genome20, 21. This matter is further complicated by the fact that MGEs are themselves often vectors of HGT, as we will explain below.
Molecular mechanisms for prokaryotic HGT have been established in many experimental systems, beginning with the discovery of pneumococcal capsule acquisition22. Overall, three major categories have been studied extensively: conjugation, transduction, and natural transformation. Conjugation involves the direct transfer of DNA from a donor to a recipient cell during physical contact through pili23 or pore-like structures24. It is typically orchestrated by conjugative plasmids or transposons which carry the necessary genetic functions to ensure their own transfer via this process. Transduction refers to the transfer of non-viral DNA encapsulated in virus or virus-like particles. During lytic infections, the viruses of bacteria (known as phages) can package parts of the lysing host’s genome into a small proportion of their particles, and this DNA may then be delivered to distant cells25, 26. Natural transformation involves the uptake of free DNA from the environment, occurring after the recipient cell enters a physiological state known as ‘competence’27. Although natural transformation of competent bacteria is a host-encoded process and usually results in unbiased DNA uptake, most known examples of conjugation and transduction are tied to the activity of MGEs that need not benefit the host and can even be detrimental (Box 2). Hence, molecular barriers which limit the spread of MGEs can directly contribute to prokaryotic survival12, 28, 29. Among these defense mechanisms, the R–M and CRISPR–Cas systems are unique in that they can eliminate diverse MGEs without forfeiting selectivity, and thus generally permit HGT while minimizing the risk of genomic parasitism. These features provide a central basis for comparison to mammalian immunity, which, we argue, is analogous in its ability to tolerate diverse microbiota while safeguarding against disease-causing pathogens.
Box 2. Mobile genetic elements and the threat of genomic parasitism.
Unlike typical genes which rely strictly on a host genome for carriage, mobile genetic elements (MGEs) can propagate independently of host replication. Therefore, their presence in genomes could be the result of parasitism rather than natural selection for their phenotypic contributions to host survival20. For example, although conjugative plasmids and transposons can carry adaptive traits, such as genes for antibiotic resistance which promote survival, horizontal dissemination of these elements can occur even when they burden their hosts with extraneous genetic cargoes that do not promote fitness or vertical transmission. Lytic phages represent an extreme form of genomic parasitism in that they immediately dispose of their host after the process of self-amplification. On the other hand, the ‘temperate’ phages offer some apparent flexibility, and can partake in an alternative infection state that spares the cell from lysis, known as lysogeny77. Lysogeny often involves integration into the bacterial chromosome as a so-called prophage, where the host can tend to its replication. Temperate phages can thus alter a recipient genome directly through lysogenization140–142, but this is not necessarily a stable arrangement either: Functional prophages retain the ability to re-initiate the lytic cycle and excise from the chromosome, both stochastically and in response to DNA damage or other signals that their host is compromised78, 143, 144.
Resistance and tolerance within prokaryotes
Whereas microbiome analyses have uncovered substantial metagenomic variation among multicellular organisms within a species (Box 1), comparative genome analyses of prokaryotes have revealed a surprising degree of genomic variability among related strains which were traditionally classified as members of the same species30, 31. Accordingly, a modern view of prokaryotic genomes has emerged which distinguishes ‘core’ genome sequences common to most or all strains of a particular group from the ‘accessory’ genome sequences that are not universally sampled32, 33. Accessory sequences typically comprise no more than 10–20% of a given genome34, but their genic diversity across strains can be substantially greater than that observed for core sequences18. Furthermore, it has been found that prophages, plasmids, and various predicted MGEs are usually associated with accessory rather than core sequences of the genome35–39. Thus, by analogy to mammal-associated microbiomes, the accessory genomes of prokaryotes may represent a transient repository for horizontally derived foreign genetic information which can contribute to adaptability. However, since both microbiomes and prokaryotic accessory genomes are also liable to harbour parasitic elements, the host organism may employ its selective defenses to keep these elements in check whilst participating in symbiotic interactions or HGT. Within prokaryotic hosts, the resistance and tolerance capacities of R–M or CRISPR–Cas systems are well-suited to fulfill this task.
Restriction–modification systems
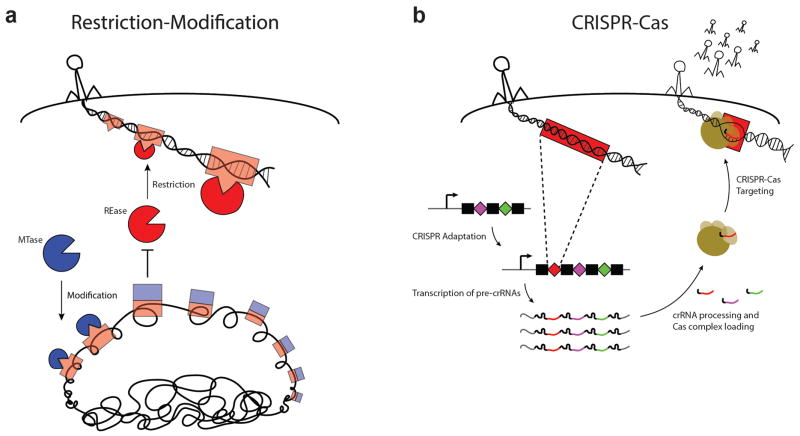
A minimal R–M system encodes enzymes with two activities: a restriction endonuclease (REase) which cleaves double-stranded DNA upon recognition of specific target sequences, and a methyltransferase (MTase) which modifies these sites via methylation to prevent cleavage4. These activities may be associated with separate proteins, a complex, or a single protein, depending on the type of system (for a comprehensive review on their nomenclature, see:40). Recognition sites for an R–M system are typically palindromic sequences of 4–8 bp in length — short enough to occur frequently in the genome of a prokaryotic organism at random. Hence, without a nuclear envelope to insulate their genetic material from the cytoplasm, prokaryotes employ methylation to protect their chromosome and other native DNA elements from attack by the REase. Meanwhile, the REase can provide immunity to invading genetic elements which have not had sufficient exposure to the cell’s MTase, including the DNA of phages and plasmids41 (Fig. 2a). Thus, methylation provides the R–M system with an intrinsic basis for distinguishing ‘self’ from ‘non-self’; and, similar to pattern recognition by mammalian pattern recognition receptors3, each system is hardwired for surveillance of a particular recognition sequence. In these regards, R–M activities are akin to mammalian innate immune functions, and may serve an analogous role in the resistance against viruses and other MGEs (but see 42 for a review on additional functions that have been described for some of these systems).
Figure 2. Prokaryotic analogs of innate and adaptive immunity.
a | Antiviral defense mediated by Restriction-Modification systems. Restriction endonuclease enzymes (“REase,” red) cleave invading viral DNA at short sequence motifs known as recognition sites (red boxes). Methyltransferase enzymes (“MTase,” blue) can modify DNA at the same recognition site sequences (appended with blue boxes), in order to prevent cleavage by their cognate REase. The sequence-specificity of a REase is hardwired for a particular recognition site, and thereby offers innate immunity to unmodified viruses that harbor these sites in their DNA. While unmodified invading DNA of mobile genetic elements is rarely methylated fast enough to receive protection from restriction, modification is generally effective in preventing cleavage of the host chromosome and thus allows for a rudimentary form of self/non-self discrimination. b | Antiviral defense mediated by CRISPR–Cas systems. CRISPR arrays are composed of alternating units of repeat sequences (black rectangles) interrupted by unique spacer sequences (colored diamonds). Newly encountered phage sequences (red) can be incorporated as spacer DNA within the host’s CRISPR array through the process of CRISPR adaptation, providing a genetic memory of past infection. Transcription of the CRISPR array provides primary transcripts (pre-crRNAs) that are processed into short, mature species which each include a single spacer sequence. During CRISPR–Cas targeting, Cas protein complexes are guided by individual mature crRNAs to mediate the destruction of invading nucleic acids that harbor a matching target sequence. By virtue of sequences in their flanking repeat elements, spacer DNA of the CRISPR array is intrinsically spared from CRISPR–Cas targeting in order to prevent autoimmunity50, 60.
R–M systems have been identified in about 75% of sequenced prokaryotes, averaging roughly two systems per genome43. As we alluded above, the relative ubiquity of their recognition sequences in DNA dictates that an R–M system’s selectivity for foreign target elements must be primarily informed by methylation patterns (although underrepresentation of recognition sequences can also occur44). However, this rudimentary mechanism for self-/non-self-discrimination is particularly susceptible to host mimicry evasion paradigms45; even exogenous DNA may bypass restriction if it arrives pre-methylated (for example, via modification in a neighboring cell possessing the same MTase)46, 47. In this scenario, the methylated foreign DNA is effectively tolerated as a self component and recognized by the host’s MTase during subsequent rounds of replication. Therefore, an infectious MGE can propagate freely in spite of the population’s REase surveillance once it manages to achieve methylation. In parallel, prokaryotic organisms may acquire resistance to these infectious elements through other defense mechanisms, such as their CRISPR–Cas systems48.
CRISPR–Cas systems
Three distinct types of CRISPR–Cas systems have now been defined49, but they all share two key components: a CRISPR locus comprised of clustered, regularly interspaced, short palindromic repeat sequences, and a set of genes which encode Cas (CRISPR–associated) proteins (reviewed in 50). Cas proteins include nucleases which can eliminate invading DNA targets, and they complex with small RNA guides known as CRISPR RNAs (crRNAs) to identify their targets via base pair complementarity51–53. crRNAs are derived from transcription of the host’s CRISPR locus, which is structured as an array of short, palindromic repeat sequences (~40 bp in length) that are intercalated with unique sequences of a similar length, known as spacers. After being processed into a small, mature species, crRNAs contain a single spacer sequence which specifies the target for its Cas nuclease complex51, 54. In this way, Cas nucleases can be programmed to recognize many different DNA targets, according to the spacer sequence of their crRNA. Importantly, the spacer content of a cell’s CRISPR locus can be actively modified through a process known as CRISPR adaptation55 (see 56 for a review). During this process, spacer DNA appears to be taken directly from an invading element for incorporation into the CRISPR locus (Fig. 2b). This provides the cell with a novel target sequence for its Cas nucleases, as well as a genetic memory of the encounter to pass on to daughter cells. In this manner, CRISPR–Cas function is analogous to mammalian adaptive immunity. However, it should be noted that memory acquired through CRISPR is fully heritable, while newborn mammals only receive short-lived, maternally derived passive immunity57. Furthermore, whereas the mammalian immune system relies heavily on clonal deletion and anergization of self-reactive cells to establish central tolerance58, CRISPR–Cas systems do not require such mechanisms since spacer sequence diversity is not randomly generated, but rather derived from invading genetic elements in an apparent Lamarckian evolutionary fashion6, 59. Nevertheless, because the spacer sequences in CRISPR loci are themselves perfect matches for the crRNAs they encode, additional mechanisms are required to protect the CRISPR locus DNA from autoimmune responses50, 60. In this sense, CRISPR loci can be viewed as immune-privileged regions of the genome.
CRISPR–Cas systems are found in 90% of archaeal genomes and 50% of sequenced bacteria61. The majority of naturally occurring spacer sequences with known targets match to viral elements, although matches to plasmids and other MGEs are also observed62. Furthermore, a growing body of experimental work indicates that CRISPR–Cas systems are capable of eliminating a wide range of MGEs targeted by their spacers55, 63, 64. Unlike R–M systems, which discriminate targets primarily via methylation, CRISPR–Cas systems derive their selectivity first and foremost from an exquisitely specific sequence recognition capability based on crRNA complementarity. The spacer sequence of a typical mature crRNA is 20–40 nucleotides — long enough to discriminate different target elements on the basis of sequence alone. Likewise, a phage- or plasmid-derived spacer sequence is unlikely to specify targeting of the host’s own chromosome (generally not exceeding ~5 Mbp for a prokaryotic genome18). On the other hand, it has been shown that spacers engineered to specifically target the chromosome are indeed lethally self-reactive65, 66, so the observation that self-targeting spacers rarely occur in nature could be explained by the immediate culling of cells which acquire such spacers — in a manner akin to clonal deletion. Interestingly, however, experimental evidence indicates that spacer acquisition is biased towards extra-chromosomal sequences from the outset (that is, even in the absence of CRISPR–Cas target degradation), despite the overrepresentation of chromosomal DNA by mass67, 68. This bias helps to ensure that CRISPR–Cas surveillance displays a selective preference for foreign genetic elements, since, only a subset of HGT events result in physical linkage of DNA to the chromosome.
Perspectives on tolerance
The flux of genetic information in prokaryotic genomes, facilitated by HGT, can be selectively moderated by both CRISPR–Cas and R–M systems through their influence on MGEs and accessory genomic content. Although the selectivity of these systems is tied to their resistance mechanisms, it can be reinforced by tolerance in certain contexts. As outlined above, a CRISPR–Cas system will resist diverse MGEs according to its spacer content, and this can have evolutionary consequences for lineages that acquire different spacers. For example, cells which acquire immunity to parasitic elements can gain a selective advantage55, while those that target favorable elements can be put at a disadvantage and eventually be lost from the population63, 69, 70. However, just as the risk imposed by particular microbes can be niche- or context-dependent within a mammalian host71–73, the fitness contributions associated with a particular MGE are not always clear-cut in the prokaryotic domain. Temperate phages are a prime example (Box 2). Although toxic during lytic infections, they can be maintained as prophages in an alternative, lysogenic state which does not necessarily reduce their host’s fitness and can even be advantageous74–76. Notwithstanding, indiscriminate CRISPR–Cas targeting of temperate phage DNA will compromise the stability of the lysogenic state in addition to preventing lytic infection64. This is because the transition between the two states does not involve genetic alteration of the phage’s DNA sequence77 but rather results from changes in its transcriptional activity within the host; in fact, most of the prophage genes are repressed during lysogeny78.
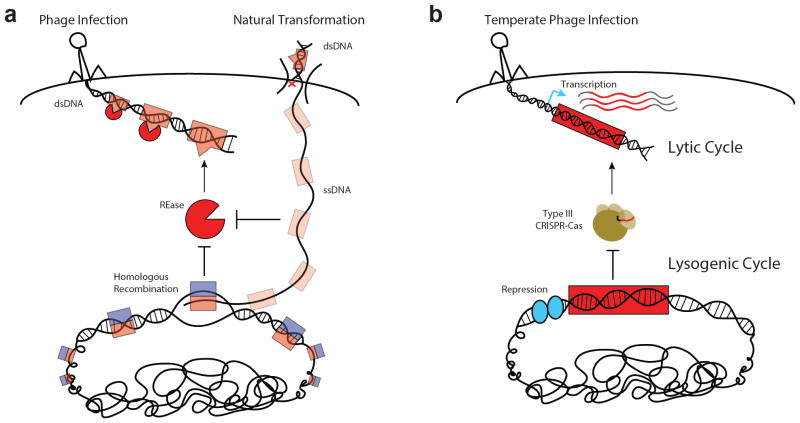
Recently, we showed that this property allows a staphylococcal type III CRISPR–Cas system to distinguish between each infection state (that is, lytic versus lysogenic) via transcription-dependent targeting79. Type III CRISPR–Cas systems only initiate an immune response when their target sequences are transcribed. As such, they can tolerate non-transcribed prophage targets during lysogeny without withdrawing resistance to lytic infection by the same phage (FIG. 3b). Tolerance, at least for temperate phages, may thus be employed in a context-dependent, conditional manner, as it is for microbiota in the gut of a healthy mammal80. This prokaryotic phenomenon could be further viewed as a ‘disease tolerance’ paradigm81–83, insofar as it averts the ‘immunopathology’ associated with targeting of prophages in the host chromosome without elimination of the target element. Alternative strategies for disease tolerance, aimed at neutralizing parasite-derived toxins84, 85 rather than the parasites themselves, might also exist in the prokaryotic domain. Past and recent work have demonstrated that type III CRISPR–Cas systems can cleave RNA targets both in vitro86–89 and in vivo89–91, and thus offer the potential to reduce toxicity associated with certain viral transcripts. If proven to exist, type III systems that can cleave RNA without degrading DNA could offer another avenue for tolerating MGEs. Current evidence, however, suggests that RNA cleavage is not sufficient to rescue cells from lysis by DNA viruses92.
Figure 3. Tolerance of foreign DNA during prokaryotic nucleic acid surveillance.
a | REases can recognize and inactivate double-stranded DNA (dsDNA) of unmodified phages and other MGEs, but do not recognize single-stranded DNA (ssDNA) intermediates of natural transformation. Successful recombination may result from pairing of unmethylated ssDNA with a methylated homologous sequence in the host chromosome, which promotes rapid modification and tolerance of the newly incorporated strand. Red ‘x’ represents arbitrary degradation of a donor DNA strand upon uptake of dsDNA during natural transformation. b | Type III CRISPR–Cas systems can only attack DNA elements with an actively transcribed target sequence. During a temperate phage lytic cycle, most sequences of the phage genome are transcribed and may license targeting by a type III CRISPR–Cas system. During its lysogenic cycle, most prophage sequences are repressed in the host chromosome and the prophage is spared from type III targeting.
In certain contexts, R–M systems can also tolerate insertion of foreign DNA into the chromosome. Owing to their intracellular, double-strand cleavage mechanism, REases do not eliminate the DNA of unmodified target elements per se. In fact, it has been shown that fragments of restriction-sensitive DNA encountered through transduction or conjugation can be rescued via recombination with the chromosome93–95. This was proposed to contribute to the genomic mosaicism observed among natural isolates of E. coli96. Furthermore, evidence indicates that R–M systems are ineffective at blocking natural transformation with otherwise restriction-sensitive DNA, at least when homologous sequences are introduced97–101. These results have been explained in light of the findings that DNA enters the cell through a single-stranded intermediate during natural transformation102, 103, which remains stable104–106 prior to recombination with a methylated complementary strand (FIG. 3a). Interestingly, an R–M system in S. pneumoniae was found to encode an auxiliary MTase which is upregulated during competence and allows even non-homologous sequences to be protected from restriction during natural transformation107, 108. Known as DpnA, this MTase preferentially methylates ssDNA, and may thus promote natural transformation without compromising the system’s ability to resist phages which enter double stranded. In its absence, replication of non-homologous, unmethylated ssDNA which has integrated into the chromosome can give rise to unmethylated dsDNA that is susceptible to the REase109. Hence, we suggest that the specialized action of DpnA exemplifies another disease tolerance strategy in that it is only required to protect the chromosome from immunopathological damage when foreign, non-homologous sequences are introduced. Non-homologous fragments of DNA that are introduced via phage transduction might also be processed to single strands prior to recombination. The effect of DpnA on transduction efficiency from an unmodified donor should therefore be examined in future work. Finally, it should be emphasized that the tolerance scenario observed with DpnA closely mirrors that observed with the type III CRISPR–Cas system, in which a substantial stretch of non-homologous DNA — the prophage — is allowed to integrate into the chromosome.
Concluding remarks
Surveillance by both R–M and CRISPR–Cas systems can be optimized for selective incorporation of foreign genetic information that reduces the risks of parasitism and immunopathology. This is similar to the pattern observed for multicellular organisms, where an optimal balance of resistance and tolerance must be struck to accommodate commensal microbiota without succumbing to infection or compromising health110. Mammalian strategies for pathogen resistance have been studied extensively in the field of immunology. Meanwhile, host strategies for tolerance of non-self elements, especially as an alternative or at least auxiliary immune function with respect to resistance, have been explored far less in animal models110–112. Efforts to understand tolerance mechanisms hold the promise of revitalizing our grasp on clinical problems in cases where vaccination, antibiotics, and other therapeutic interventions aimed at bolstering resistance have fallen short. Alternative therapeutic approaches could be aimed at exploiting or reinforcing tolerance83. Of particular relevance to our discussion, it has been postulated that tolerance strategies can lead to more stable (or at least more homogeneous) evolutionary outcomes113–115 that are distinct from the arms race dynamics which typically result from resistance to pathogens (for further discussion of the latter, see:116). The outcomes observed for CRISPR–Cas targeting of temperate phages in prokaryotes are ostensibly consistent with this notion, as tolerance during lysogeny does not lead to genetic alteration of either the host or its target prophage. In the absence of tolerance, lysogenization was less frequent and only occurred in genetic mutants which had either lost their prophage target sequence or dismantled their CRISPR–Cas immunity79. We speculate that this latter propensity to abandon CRISPR–Cas immunity (see also: 69, 70, 117, 118) might in part explain the observed absence of these systems in about half of sequenced bacteria, especially considering that mechanisms for non-self tolerance have not been identified in the other two CRISPR–Cas branches. In other words, fitness costs associated with CRISPR–Cas immunopathology and/or resistance of favorable non-self elements could drive their loss from prokaryotic genomes in the context of population bottlenecks. Analogous complications faced by mammalian lineages are dealt with through well-conserved central and peripheral tolerance mechanisms. The latter mechanisms appear to be particularly critical during encounters with innocuous non-self elements, including food-derived products and commensal microbes that are highly abundant in the gut119. Insofar as tolerance strategies work to curb — or even replace — resistance strategies that would otherwise reduce a particular microbial burden, tolerance will, in turn, influence microbiome compositions and stability. Therefore, in addition to leading us towards novel therapeutics for dealing with the complications of infection and immunopathology83, knowledge of tolerance mechanisms that fulfill these criteria could prove insightful when attempting to manipulate commensal members of the microbiota for therapeutic purposes.
Acknowledgments
We would like to thank Daniel Mucida and Philip M. Nussenzweig at The Rockefeller University for critical discussion and corrections to the manuscript. L.A.M is supported by the Searle Scholars Program, Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01).
References
- 1.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–78. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 2.Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–72. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–26. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 4.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr Opin Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Goren M, Yosef I, Edgar R, Qimron U. The bacterial CRISPR/Cas system as analog of the mammalian adaptive immune system. RNA Biol. 2012;9:549–554. doi: 10.4161/rna.20177. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R, Marraffini LA. CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller HJ. Some Genetic Aspects of Sex. The American Naturalist. 1932;66:118–138. [Google Scholar]
- 9.Muller HJ. The Relation of Recombination to Mutational Advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 10.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–56. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–92. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 12.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 13.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–18. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 14.Dunning Hotopp JC. Horizontal gene transfer between bacteria and animals. Trends in Genetics. 2011;27:157–163. doi: 10.1016/j.tig.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersson JO, Doolittle WF, Nesbo CL. Genomics. Are there bugs in our genome? Science. 2001;292:1848–50. doi: 10.1126/science.1062241. [DOI] [PubMed] [Google Scholar]
- 16.Takeuchi N, Kaneko K, Koonin EV. Horizontal gene transfer can rescue prokaryotes from Muller’s ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 2014;4:325–39. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414–24. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 18.Koonin EV, Wolf YI. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 2008;36:6688–719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–7. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 21.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–3. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 22.Griffith F. The Significance of Pneumococcal Types. J Hyg (Lond) 1928;27:113–59. doi: 10.1017/s0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 24.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zinder ND, Lederberg J. Genetic exchange in Salmonella. J Bacteriol. 1952;64:679–99. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinstock GM. Modern Microbial Genetics. John Wiley & Sons, Inc; 2002. pp. 561–579. [Google Scholar]
- 27.Johnston C, Martin B, Fichant G, Polard P, Claverys JP. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat Rev Microbiol. 2014;12:181–96. doi: 10.1038/nrmicro3199. [DOI] [PubMed] [Google Scholar]
- 28.Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–327. doi: 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 29.Makarova KS, Wolf YI, Koonin EV. Comparative genomics of defense systems in archaea and bacteria. Nucleic Acids Res. 2013;41:4360–77. doi: 10.1093/nar/gkt157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray AE, et al. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc Natl Acad Sci U S A. 2001;98:9853–8. doi: 10.1073/pnas.171178898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–5. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan R, Reeves PR. Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol. 2000;8:396–401. doi: 10.1016/s0966-842x(00)01791-1. [DOI] [PubMed] [Google Scholar]
- 33.Mira A, Martin-Cuadrado AB, D’Auria G, Rodriguez-Valera F. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 34.Makarova KS, et al. Dark matter in archaeal genomes: a rich source of novel mobile elements, defense systems and secretory complexes. Extremophiles. 2014;18:877–93. doi: 10.1007/s00792-014-0672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindsay JA, Holden MT. Understanding the rise of the superbug: investigation of the evolution and genomic variation of Staphylococcus aureus. Funct Integr Genomics. 2006;6:186–201. doi: 10.1007/s10142-005-0019-7. [DOI] [PubMed] [Google Scholar]
- 36.Cortez D, Forterre P, Gribaldo S. A hidden reservoir of integrative elements is the major source of recently acquired foreign genes and ORFans in archaeal and bacterial genomes. Genome Biol. 2009;10:R65. doi: 10.1186/gb-2009-10-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R. Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 2011;11:224. doi: 10.1186/1471-2180-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Bakker HC, et al. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS One. 2013;8:e67511. doi: 10.1371/journal.pone.0067511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozer EA, Allen JP, Hauser AR. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics. 2014;15:737. doi: 10.1186/1471-2164-15-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts RJ, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–12. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arber W, Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 42.Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliveira PH, Touchon M, Rocha EP. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014;42:10618–31. doi: 10.1093/nar/gku734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocha EP, Danchin A, Viari A. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 2001;11:946–58. doi: 10.1101/gr.gr-1531rr. [DOI] [PubMed] [Google Scholar]
- 45.Damian RT. Molecular Mimicry: Antigen Sharing by Parasite and Host and Its Consequences. The American Naturalist. 1964;98:129–149. [Google Scholar]
- 46.Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J Bacteriol. 1953;65:113–21. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korona R, Levin BR. Phage-Mediated Selection and the Evolution and Maintenance of Restriction-Modification. Evolution. 1993;47:556–575. doi: 10.1111/j.1558-5646.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- 48.Dupuis ME, Villion M, Magadan AH, Moineau S. CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat Commun. 2013;4:2087. doi: 10.1038/ncomms3087. [DOI] [PubMed] [Google Scholar]
- 49.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 51.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 56.Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Molecular Microbiology. 2014;93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grindstaff JL, Brodie ED, 3rd, Ketterson ED. Immune function across generations: integrating mechanism and evolutionary process in maternal antibody transmission. Proc Biol Sci. 2003;270:2309–19. doi: 10.1098/rspb.2003.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 59.Haerter JO, Sneppen K. Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus. mBio. 2012;3:e00126–12. doi: 10.1128/mBio.00126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodt A, Lurie-Weinberger MN, Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biology direct. 2011;6:65. doi: 10.1186/1745-6150-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R, Qimron U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bikard D, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–50. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–10. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 70.Jiang W, et al. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 2013;9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal Carriage as a Source of Staphylococcus aureus Bacteremia. New England Journal of Medicine. 2001;344:11–16. doi: 10.1056/NEJM200101043440102. [DOI] [PubMed] [Google Scholar]
- 72.Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans. Curr Opin Microbiol. 2004;7:336–41. doi: 10.1016/j.mib.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Stecher B, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–89. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edlin G, Lin L, Bitner R. Reproductive fitness of P1, P2, and Mu lysogens of Escherichia coli. J Virol. 1977;21:560–4. doi: 10.1128/jvi.21.2.560-564.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dykhuizen D, Campbell JH, Rolfe BG. The influences of a λ prophage on the growth rate of Escherichia coli. Microbios. 1978;23:99–113. [PubMed] [Google Scholar]
- 76.Brüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lwoff A. Lysogeny. Bacteriol Rev. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Johnson AD, et al. λ Repressor and cro—components of an efficient molecular switch. Nature. 1981;294:217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- 79.Goldberg GW, Jiang W, Bikard D, Marraffini LA. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature. 2014;514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gozzelino R, et al. Metabolic adaptation to tissue iron overload confers tolerance to malaria. Cell Host Microbe. 2012;12:693–704. doi: 10.1016/j.chom.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 82.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335:936–41. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soares MP, Gozzelino R, Weis S. Tissue damage control in disease tolerance. Trends Immunol. 2014;35:483–94. doi: 10.1016/j.it.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 84.Playfair JH, Taverne J, Bate CA, de Souza JB. The malaria vaccine: anti-parasite or anti-disease? Immunol Today. 1990;11:25–7. doi: 10.1016/0167-5699(90)90007-v. [DOI] [PubMed] [Google Scholar]
- 85.Schofield L, Hewitt MC, Evans K, Siomos MA, Seeberger PH. Synthetic GPI as a candidate anti-toxic vaccine in a model of malaria. Nature. 2002;418:785–9. doi: 10.1038/nature00937. [DOI] [PubMed] [Google Scholar]
- 86.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, et al. Structure and Mechanism of the CMR Complex for CRISPR-Mediated Antiviral Immunity. Mol Cell. 2012;45:303–313. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Staals RH, et al. RNA Targeting by the Type III-A CRISPR-Cas Csm Complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamulaitis G, et al. Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- 90.Hale CR, et al. Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–8. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Samai P, et al. Co-transcriptional DNA and RNA Cleavage during Type III CRISPR-Cas Immunity. Cell. 2015;161:1164–74. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wood WB. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966;16:118–33. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- 94.McKane M, Milkman R. Transduction, restriction and recombination patterns in Escherichia coli. Genetics. 1995;139:35–43. doi: 10.1093/genetics/139.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Milkman R, et al. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics. 1999;153:539–54. doi: 10.1093/genetics/153.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Milkman R, Bridges MM. Molecular evolution of the Escherichia coli chromosome. IV. Sequence comparisons. Genetics. 1993;133:455–68. doi: 10.1093/genetics/133.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Trautner TA, Pawlek B, Bron S, Anagnostopoulos C. Restriction and modification in B. subtilis. Biological aspects. Mol Gen Genet. 1974;131:181–91. doi: 10.1007/BF00267958. [DOI] [PubMed] [Google Scholar]
- 98.Harris-Warrick RM, Lederberg J. Interspecies transformation in Bacillus: sequence heterology as the major barrier. J Bacteriol. 1978;133:1237–45. doi: 10.1128/jb.133.3.1237-1245.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bron S, Luxen E, Trautner TA. Restriction and modification in B. subtilis: the role of homology between donor and recipient DNA in transformation and transfection. Mol Gen Genet. 1980;179:111–7. doi: 10.1007/BF00268452. [DOI] [PubMed] [Google Scholar]
- 100.Lacks SA, Springhorn SS. Transfer of recombinant plasmids containing the gene for DpnII DNA methylase into strains of Streptococcus pneumoniae that produce DpnI or DpnII restriction endonucleases. J Bacteriol. 1984;158:905–9. doi: 10.1128/jb.158.3.905-909.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cohan FM, Roberts MS, King EC. The Potential for Genetic Exchange by Transformation within a Natural Population of Bacillus subtilis. Evolution. 1991;45:1393–1421. doi: 10.1111/j.1558-5646.1991.tb02644.x. [DOI] [PubMed] [Google Scholar]
- 102.Lacks S. Molecular fate of DNA in genetic transformation of Pneumococcus. J Mol Biol. 1962;5:119–31. doi: 10.1016/s0022-2836(62)80067-9. [DOI] [PubMed] [Google Scholar]
- 103.Piechowska M, Fox MS. Fate of transforming deoxyribonucleate in Bacillus subtilis. J Bacteriol. 1971;108:680–9. doi: 10.1128/jb.108.2.680-689.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eisenstadt E, Lange R, Willecke K. Competent Bacillus subtilis cultures synthesize a denatured DNA binding activity. Proc Natl Acad Sci U S A. 1975;72:323–7. doi: 10.1073/pnas.72.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morrison DA, Mannarelli B. Transformation in pneumococcus: nuclease resistance of deoxyribonucleic acid in the eclipse complex. J Bacteriol. 1979;140:655–65. doi: 10.1128/jb.140.2.655-665.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mortier-Barriere I, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–36. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 107.Cerritelli S, Springhorn SS, Lacks SA. DpnA, a methylase for single-strand DNA in the Dpn II restriction system, and its biological function. Proc Natl Acad Sci U S A. 1989;86:9223–7. doi: 10.1073/pnas.86.23.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Johnston C, Martin B, Granadel C, Polard P, Claverys JP. Programmed protection of foreign DNA from restriction allows pathogenicity island exchange during pneumococcal transformation. PLoS Pathog. 2013;9:e1003178. doi: 10.1371/journal.ppat.1003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Johnston C, Martin B, Polard P, Claverys JP. Postreplication targeting of transformants by bacterial immune systems? Trends Microbiol. 2013;21:516–21. doi: 10.1016/j.tim.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 110.Ayres JS, Schneider DS. Tolerance of infections. Annu Rev Immunol. 2012;30:271–94. doi: 10.1146/annurev-immunol-020711-075030. [DOI] [PubMed] [Google Scholar]
- 111.Raberg L, Graham AL, Read AF. Decomposing health: tolerance and resistance to parasites in animals. Philos Trans R Soc Lond B Biol Sci. 2009;364:37–49. doi: 10.1098/rstb.2008.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Howick VM, Lazzaro BP. Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evol Biol. 2014;14:56. doi: 10.1186/1471-2148-14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schafer JF. Tolerance to Plant Disease. Annual Review of Phytopathology. 1971;9:235–252. [Google Scholar]
- 114.Roy BA, Kirchner JW. Evolutionary dynamics of pathogen resistance and tolerance. Evolution. 2000;54:51–63. doi: 10.1111/j.0014-3820.2000.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 115.Miller MR, White A, Boots M. The evolution of parasites in response to tolerance in their hosts: the good, the bad, and apparent commensalism. Evolution. 2006;60:945–56. [PubMed] [Google Scholar]
- 116.Brockhurst MA, Koskella B. Experimental coevolution of species interactions. Trends Ecol Evol. 2013;28:367–75. doi: 10.1016/j.tree.2013.02.009. [DOI] [PubMed] [Google Scholar]
- 117.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci USA. 2011;108:20136–20141. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vercoe RB, et al. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5:232–9. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99:15451–5. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Backhed F, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 123.Amaral FA, et al. Commensal microbiota is fundamental for the development of inflammatory pain. Proc Natl Acad Sci U S A. 2008;105:2193–7. doi: 10.1073/pnas.0711891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci U S A. 2008;105:2117–22. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bruls T, Weissenbach J. The human metagenome: our other genome? Hum Mol Genet. 2011;20:R142–8. doi: 10.1093/hmg/ddr353. [DOI] [PubMed] [Google Scholar]
- 126.Robinson CJ, Bohannan BJ, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74:453–76. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Human Microbiome Project C. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 134.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 135.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao L. Genomics: The tale of our other genome. Nature. 2010;465:879–80. doi: 10.1038/465879a. [DOI] [PubMed] [Google Scholar]
- 137.Luckey TD. Introduction to intestinal microecology. Am J Clin Nutr. 1972;25:1292–4. doi: 10.1093/ajcn/25.12.1292. [DOI] [PubMed] [Google Scholar]
- 138.Gonzalez A, et al. Our microbial selves: what ecology can teach us. EMBO Rep. 2011;12:775–84. doi: 10.1038/embor.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–90. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 140.Groman NB. Evidence for the active role of bacteriophage in the conversion of nontoxigenic Corynebacterium diphtheriae to toxin production. J Bacteriol. 1955;69:9–15. doi: 10.1128/jb.69.1.9-15.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Canchaya C, Proux C, Fournous G, Bruttin A, Brussow H. Prophage genomics. Microbiol Mol Biol Rev. 2003;67:238–76. doi: 10.1128/MMBR.67.2.238-276.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 143.Bailone A, Levine A, Devoret R. Inactivation of prophage λ repressor in vivo. J Mol Biol. 1979;131:553–72. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- 144.Schubert RA, Dodd IB, Egan JB, Shearwin KE. Cro’s role in the CI Cro bistable switch is critical for λ’s transition from lysogeny to lytic development. Genes Dev. 2007;21:2461–72. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]