Figure 1.
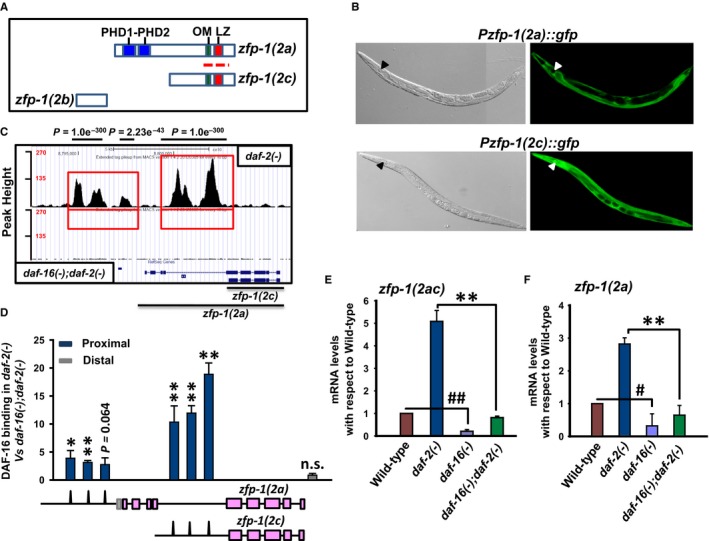
The isoforms of worm AF10 ortholog are direct transcriptional target of DAF‐16/FOXO. (A) Organization of ZFP‐1 isoforms. The polypeptide encoded by zfp‐1(2a) isoform possesses the PHD domain, OM (octapeptide motif) and LZ (leucine zipper) motifs, whereas the shorter isoform encoded by zfp‐1(2c) has the OM‐LZ motif only. The third isoform zfp‐1(2b) lacks all these domains and motifs. Red dotted line indicates region from where RNAi was designed. (B) Expression pattern of GFP driven by zfp‐1(2a) or zfp‐1(2c) promoters (right panels). Corresponding DIC images are shown in the left panels. Pharynx is marked by arrow head. (C) UCSC browser view of DAF‐16/FOXO peaks as determined by ChIP‐seq using anti‐DAF‐16/FOXO antibody (Kumar et al., 2015). Red boxes indicate the promoter regions of zfp‐1(2a) and zfp‐1(2c) where DAF‐16/FOXO peaks are observed. Lower panel shows daf‐16(mgDf50);daf‐2(e1370) [represented as daf‐16(‐);daf‐2(‐)] that lacks specific DAF‐16/FOXO peaks, while upper panel shows peaks obtained in daf‐2(e1370) [represented as daf‐2(‐)] (D) ChIP‐PCR validation of DAF‐16/FOXO binding to zfp‐1 promoters obtained by ChIP‐seq. Binding in daf‐2(‐) is normalized to that of daf‐16(‐);daf‐2(‐). The corresponding peaks on the promoters of zfp‐1, as obtained by ChIP‐seq, are shown pictographically below the graph. (E‐F) quantitative RT (QRT)–PCR detection of transcript levels for zfp‐1(2ac) or zfp‐1(2a) in WT and different mutants as mentioned. The zfp‐1(2c) transcript cannot be detected separately from zfp‐1(2a) as explained in the text. Error bars are standard deviation. **P ≤ 0.01; *P ≤ 0.05; n.s., P not significant by Student's t‐test. ## P ≤ 0.01; # P ≤ 0.05 compared to wild‐type. The graphs were plotted from three experiments.
