Figure 1.
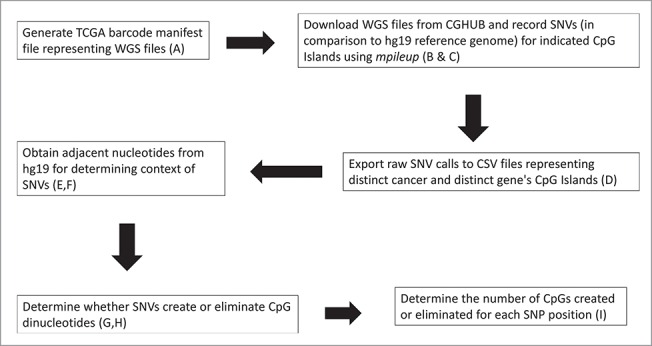
Flow chart for the processing of the BAM files from CGHub for identifying SNPs at CpG positions. (A) An example manifest file used to target a specific cancer patient's tumor DNA sequence to be downloaded from CGHub is provided in the supporting online material (SOM), entitled “Samy et al. SOM Fig. 1A, Example Manifest file.” This file is provided as an Excel file, for ease of inspection; and is provided as an XML file for direct use. Note: the Excel version of the manifest file cannot be directly used for downloads. (B) Defined CpG Island regions for each gene are included in Table 1. (C) The code used to automate the downloading and recording of variants in the patient files from TCGA is in “Samy et al. SOM Fig. 1C, Protocol_v5.” (D) The preliminary Excel files with all variants obtained from the program were combined, labeled with the TCGA cancer abbreviation, and can be found in the “Samy et al. SOM Fig. 1D, Preliminary Excel File.” Files are separated by CpG regions in the case of RB1, and the selected regions are in Table 1. (E) Adjacent nucleotides were added as a new column to the preliminary Excel files (present in SOM file labeled, “Samy et al. SOM Fig. 1D, Preliminary Excel File”) using the code (F) documented as “Samy et al. SOM Fig. 1F, GetContext.” (G) The PHP script used to determine how the variants in the DNA changed the CpG island structure is in “Samy et al. SOM Fig. 1G, cg.php.” (H) A brief summary of the algorithm guiding the script is in “Samy et al. SOM Fig. 1H, CG.PHP Explanation.” (I) The record of all rsNumbers (SNP designations) that can affect the specified CpG Islands from Table 1 are recorded in the Excel file “Samy et al. SOM Fig. 1I, RsNumbers and Cancer Counts.”
