Figure 5.
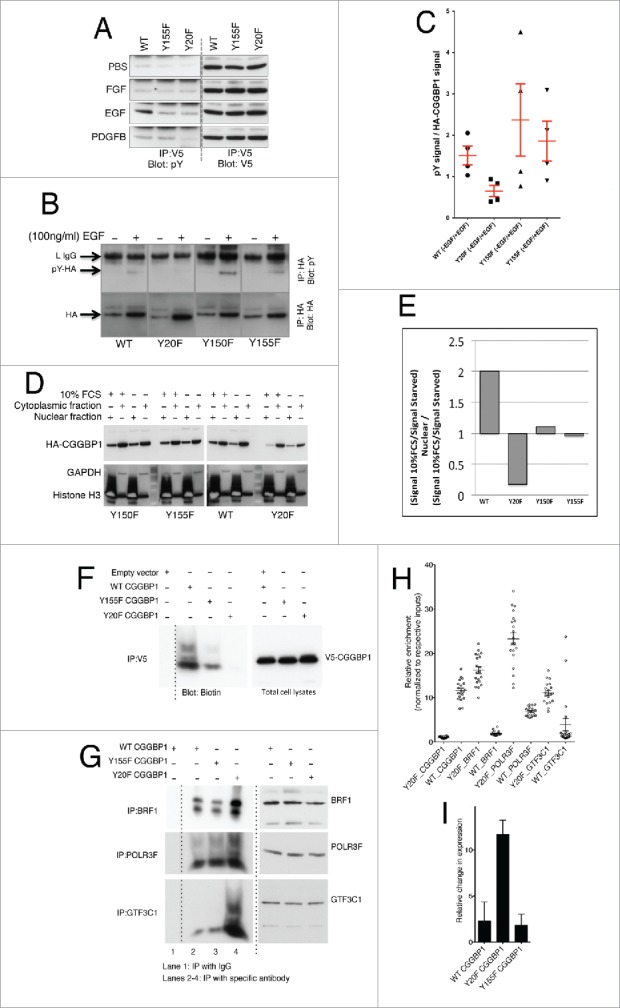
Growth stimulation induces tyrosine phosphorylation and nuclear localization of CGGBP1 that antagonizes RNA Pol III recruitment at ATE DNA. (A) 1064Sk cells transduced witAh V5-tagged WT, Y20F or Y155F CGGBP1 lentiviruses subjected to IP: V5-blot: pY after stimulation with indicated growth factors. Y20 seems to be a common phosphorylation site upon EGF and PDGFB stimulation. In all panels the upper band correspond to the light IgG chain (25 KDa approximately) and the lower band correspond to CGGBP1 (20 KDa approximately). The specificity of the IPs have been determined (not shown). (B) Measurement of the effect of EGF stimulation on tyrosine phosphorylation levels in HEK293T cells showed that Y20F mutant CGGBP1 does not respond to EGF-induced tyrosine phosphorylation. HA-tagged CGGBP1 was immunoprecipitated and blotted for pY, stripped and rebottled for HA. The HA and pY bands correspond to 20 KDa. pY signal at other molecular weights, which was stronger in EGF-treated samples, occurring due to pY-containing proteins co-immunoprecipitated with HA-CGGBP1, was ignored. (C) The net change in phosphorylation was calculated as a ratio of pY signal to total HA signal from 4 observations (WT versus Y20F; T-test p = 0.0181). (D) Consistent with immunofluorescence findings in 1064Sk cells, nuclear-cytoplasmic fractionation experiments in HEK293T cells showed that the ratio of nuclear to cytoplasmic distribution of WT CGGBP1 was increased whereas Y20F CGGBP1 was decreased upon serum stimulation; Y150F and Y155F mutations had no effect on the nuclear-cytoplasmic distribution upon serum stimulation. (E) The change in nuclear/cytoplasmic distribution upon serum stimulation was quantified using NIH ImageJ. The values are calculated using the formula ((Signal 10%FCS / Signal Quiescent) Nuclear) / ((Signal 10%FCS / Signal Quiescent) Cytoplasmic). (F) Y20 CGGBP1 exhibits strongly reduced binding to ATE DNA in in vitro DNA IPs. Equal expression of CGGBP1 and specificity of IPs have been confirmed (not shown). (G) Binding of BRF1, POLR3F and GTF3C1 were increased strongly by Y20F CGGBP1 as compared to WT CGGBP1 in in vitro DNA IPs using ATE dsDNA. For assays shown in (F) and (G), the only DNA incubated with lysates is the ATE DNA oligo pair. The smeary migration pattern on non-denaturing gel shows the mobility shift associated with interactions. (H) ChIP qPCR shows that in vivo also the binding of CGGBP1 is decreased (T-test, P < .01) but binding of BRF1, POLR3F and GTF3C1 is increased by Y20F CGGBP1 as compared to WT CGGBP1 (T-test, P < .01). (I) qPCRs show that overexpression of Y20F CGGBP1-induced Alu RNA levels compared to WT or Y155F CGGBP1-overexpression (T-test P < .0001). CGGBP1 depletion by CGGBP1 shmiR also induced Alu RNA levels compared to control shmiR (P < .0001).
