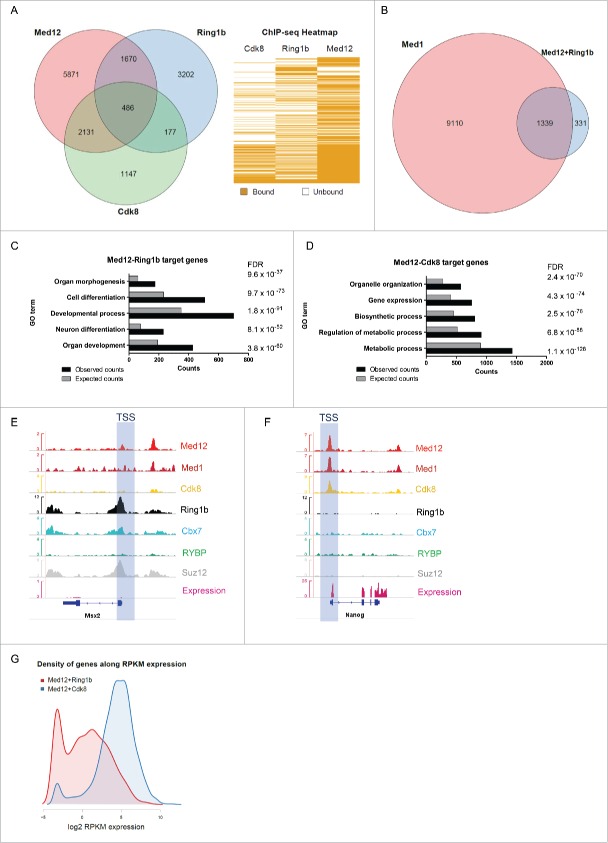
Figure 1.
Ring1b and Med12 target key developmental genes in mouse ESCs. (A) Venn diagram and heatmap of Med12, Ring1b and Cdk8 target genes in mESCs. For the Cdk8 dataset (GSE44288) we performed peak calling with a q value of 1e-5 and annotations with the closest gene promoter (+/−5 kb around the TSS). For the other data sets the qualitative occupancy data were downloaded from the supplementary material of the respective publications. (B) Venn diagram of common Ring1b-Med12 target genes (Ring1b-Med12 only-without Cdk8-, 1670 genes) that overlap with Med1 target genes. (C) GO-enriched terms of Med12-Ring1b and (D) Med12-Cdk8 target genes in mESCs. FDR represents the corrected P-value. (E-F) Med12 but not Cdk8 localize with Cbx7-PRC1 at promoters of repressed differentiation genes in mESCs. Med12 and Cdk8, but not PRC1/PRC2, localize at promoters of expressed genes in mESCs. Screenshots from ChIP-seq profiles of Med1, Med12, Cdk8, Ring1b, Cbx7, RYBP and Suz12 for selected genes. The expression levels of the selected genes (Msx2 and Nanog) are shown. (G) Density distributions of Med12+Ring1b (red) and Med12+Cdk8 (blue) target genes expression. The majority of Med12+Cdk8 targets are expressed at high levels in mESCs whereas the Med12+Ring1b targets have low or no expression. The y-axis shows the density (probability). We omitted this axis from the graph as we only intended to focus on the shift in the x-axis.