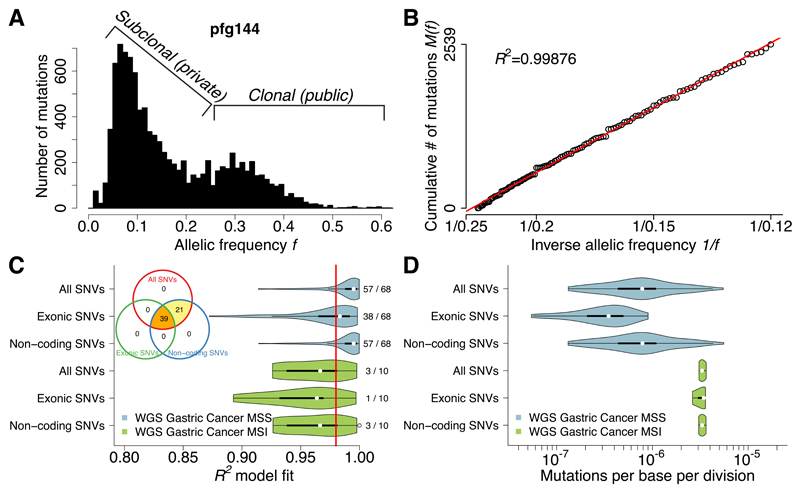
Figure 2. Neutral evolution across the whole-genome of gastric cancers.
(A) Large number of coding and non-coding mutations can be identified using WGS. (B) All detected mutations precisely accumulate as 1/f following the neutral model in this example. (C) Neutral evolution is very common in gastric cancer, with 60/78 (76.9%) samples showing goodness of fit of the neutral model R2≥0.98. This was consistent using all, exonic or non-coding subclonal mutations. The same tumors were identified as neutral by all three methods, although limitations in detecting neutrality were present when considering exonic mutations due to the limited number of variants. (D) Mutation rates were more than 4 times higher in MSI (µe=3.30×10-6) versus MSS (µe=7.82×10-7; F-test: p=1.35×10-4) cancers, consistently with the underlying biology.