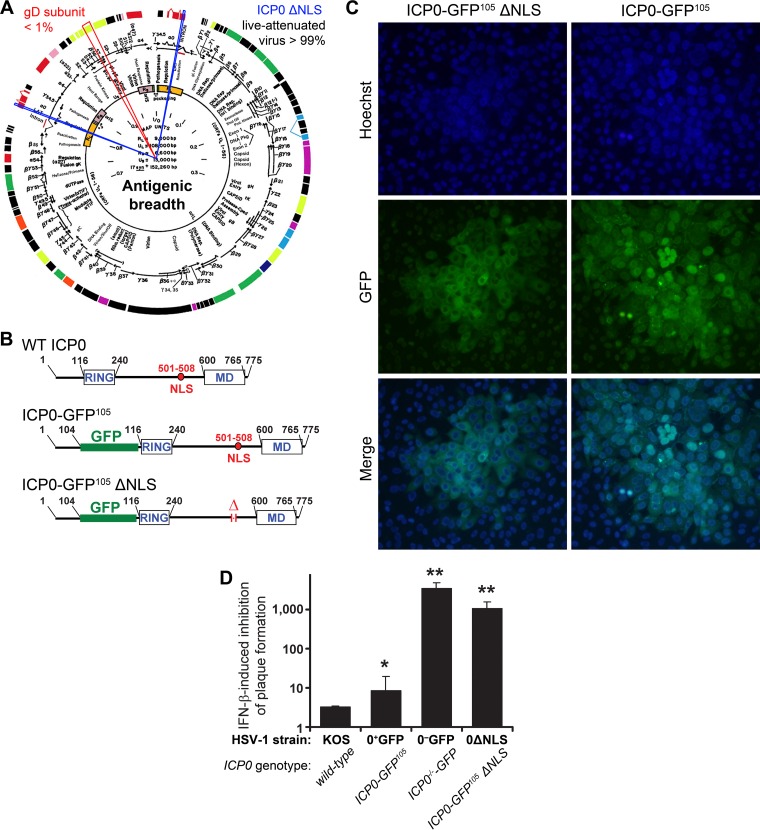
FIG 1.
Attenuated virus vaccine. (A) The ICP0 ΔNLS live attenuated HSV-1 vaccine (0ΔNLS) maintains >99% of the antigenic breadth of HSV-1 relative to the wild-type virus. In contrast, the glycoprotein D (gD) subunit vaccines tested in numerous clinical trials represents <1% of the viral proteome (red cutout). Blue cutouts on each copy of the ICP0 gene denote the loci deleted from the HSV-1 0ΔNLS vaccine strain. (B) The upper panels show a schematic of the ICP0 protein encoded by wild-type (WT) HSV-1, which includes an N-terminal RING finger motif, nuclear localization signal (NLS), and a C-terminal multimerization domain (MD). A schematic of the full-length, GFP-tagged ICP0 protein referred to here as ICP0-GFP105, which contains an insertion of the green fluorescent protein (GFP) coding sequence between codons 104 and 105 of the ICP0 gene, is shown in the middle. At bottom is a schematic of the GFP-tagged ICP0 protein encoded by HSV-1 0ΔNLS, ICP0ΔNLS-GFP105, which bears a deletion in amino acids 501 to 508 of ICP0 and removes the protein's canonical NLS (RPRKRR). (C) Comparison of single plaques of HSV-1 0ΔNLS and HSV-1 ICP0-GFP105 in a monolayer of Vero cells as photographed at 30 h postinoculation under conditions of illumination that show the nuclear dye Hoechst 33342, the GFP-tagged ICP0 proteins made by each virus (ICP0 with or without the NLS), or a merge of both. (D) Relative IFN-β sensitivity of WT HSV-1, HSV-1 ICP0-GFP105, HSV-1 0ΔNLS, or HSV-1 ICP0−/−-GFP (ICP0-null mutant) to inhibition of plaque formation in Vero cell monolayers treated with 200 U/ml IFN-β. Specifically, the column graph represents the mean ± standard deviation of the ratio of plaques that were formed when each virus was plated on ICP0-complementing L7 cells to define the actual titer versus Vero cells treated with 200 U/ml IFN-β. In prior studies, all viruses that exhibit a >500-fold inhibition of plaque formation in IFN-β-treated Vero cells have proven to be avirulent in vivo.