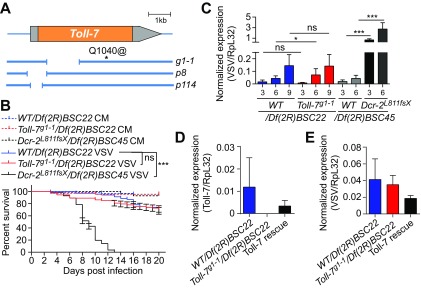
FIG 7.
Toll7g1-1 mutant flies are not sensitive to VSV infection. (A) Schematic representation of the Toll-7 gene and the deletion mutants evaluated. The coding region is highlighted in orange, and deletions are indicated by the gaps in the blue lines. For the Toll-7g1-1 allele, a stop codon replaces a glutamine at position 1040. (B) Survival of Toll-7g1-1 null mutant and control flies after VSV infection. Flies carrying a null allele of Toll-7 or controls from the same genetic background (yw; Toll-7g1-1 and y1 w1) were crossed with a line containing a deficiency covering the Toll-7 gene [Df(2R)BSC22] and hemizygous flies from the progeny were injected with conditioned medium (CM) or VSV. Dcr-2 null mutant (Dcr-2L811fsX) and WT flies crossed with the deficient Df(2R)BSC45 flies were used as positive controls. (C) Quantitative RT-PCR analysis of the accumulation of VSV RNA on the indicated day postinfection in control flies [WT/Df(2R)BSC22 and WT/Df(2R)BSC45], Toll-7 null mutant flies [Toll-7g1-1/Df(2R)BSC22], and Dcr-2 mutant flies [Dcr-2L811fsX/Df(2R)BSC45]. (D and E) Quantitative RT-PCR analysis of Toll-7 mRNA expression (D) and VSV viral RNA accumulation (E) in control [WT/Df(2R)BSC22], Toll-7 null mutant [Toll-7g1-1/Df(2R)BSC22], and Toll-7 rescue [Toll-7g1-1/Df(2R)BSC22; +/gToll-7] flies at 6 dpi. Data represent means ± standard errors (B) or SD (C, D, and E) of three independent experiments, each containing three groups of 10 (B) or 6 (C, D, and E) flies. ns, not significant; *, P < 0.05; ***, P < 0.001 (log rank [B] or unpaired t test [C, D, and E]).