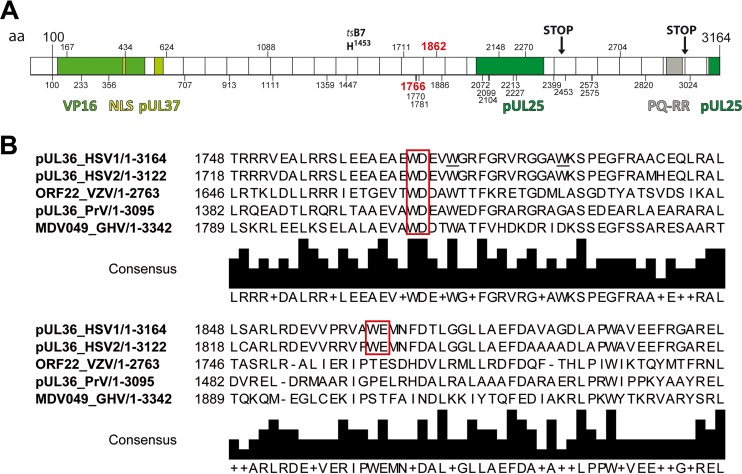
FIG 1.
Primary structure of HSV-1-pUL36. (A) Primary structure of pUL36 and location of important point mutations, truncations, interaction domains, and tryptophans. The N-terminal third of pUL36 contains an ubiquitin-specific cysteine protease (USP) with a conserved catalytic residue cysteine at position 65, a nuclear localization signal spanning residues 426 to 432, binding sites for the transcriptional activator VP16, and the inner tegument protein pUL37 (35, 41, 42, 44–46, 48, 49, 81, 109). The tsB7 point mutation Y1453H prevents genome uncoating of incoming capsids at the nuclear pores at the nonpermissive temperature (29, 110, 111). The C-terminal region of pUL36 contains two binding sites for the capsid protein pUL25 and a PQ repeat-rich region (26, 40, 112). HSV-1-pUL36 truncation mutants lacking the C-terminal 735 residues (STOP at position 2430) do not associate with capsids, whereas the 167 C-terminal residues are required during early steps of infection (STOP at position 2998) (26). The positions of 32 tryptophan residues in HSV-1-pUL36 are indicated by vertical bars and the respective numbers. W1766 and W1862 (red) were mutated in the present study. (B) Alignments around conserved W motifs. HSV-1-pUL36 was aligned with its orthologs HSV2-pUL36, VZV-ORF22, PRV-pUL36 and GHV-MDV049 using UniProtKB and Clustal Omega. The tryptophan-acidic motifs are indicated by red boxes.