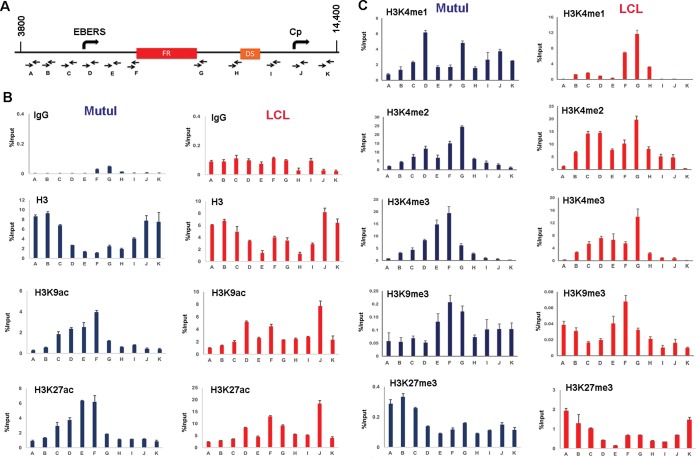
FIG 1.
Histone modifications at OriP. (A) Schematic of EBV OriP region and positions of primer sets used for ChIP-qPCR (primers A to K). (B) ChIP-qPCR of histone H3, H3K9ac, H3K27ac, or control IgG in MutuI cells (blue) or LCLs (red). (C) ChIP-qPCR of histones H3K4me1, H3K4me2, H3K4me3, H3K9me3, and H3K27me3 in MutuI cells (blue) or LCLs (red). ChIP DNA is measured as percent input, and error bars represent standard deviations for three technical replicates.