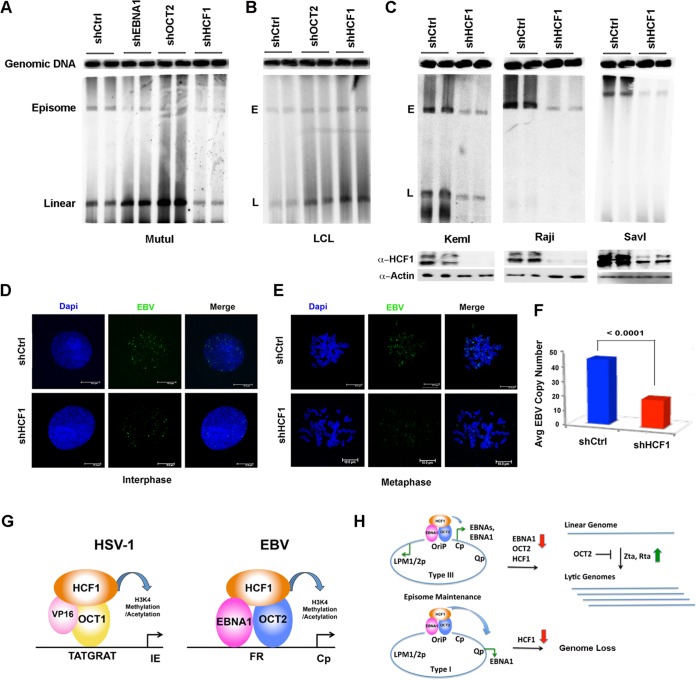
FIG 10.
Depletion of HCF1 destabilizes EBV episomes. (A) PFGE analysis of MutuI cells transduced with shCtrl, shEBNA1, shOCT2, or shHCF1. The positions of cellular genomic DNA, viral episomal (E), and linear (L) DNA are indicated. (B) PFGE analysis of LCL cells transduced with shCtrl, shOCT2, or shHCF1. (C) PFGE analysis of KemI, Raji, or SavI cells transduced with shCtrl or shHCF1. Western blots for HCF1 and actin are shown in the bottom panels. (D) FISH analysis of EBV genomes in interphase MutuI cells transduced with shCtrl or shHCF1 and counterstained with 4′,6′-diamidino-2-phenylindole (Dapi). (E) Same as in panel D, except for mitotic cells. (F) Quantification of FISH analysis shown as representatives in panels D and E. (G) Model of HCF1 function as coactivator for EBNA1-OCT2 at FR resembling the HCF1 cofactor for VP16 and OCT1 at HSV IE genes. (H) Model of HCF1 function at OriP for transcription enhancer function and episome maintenance.