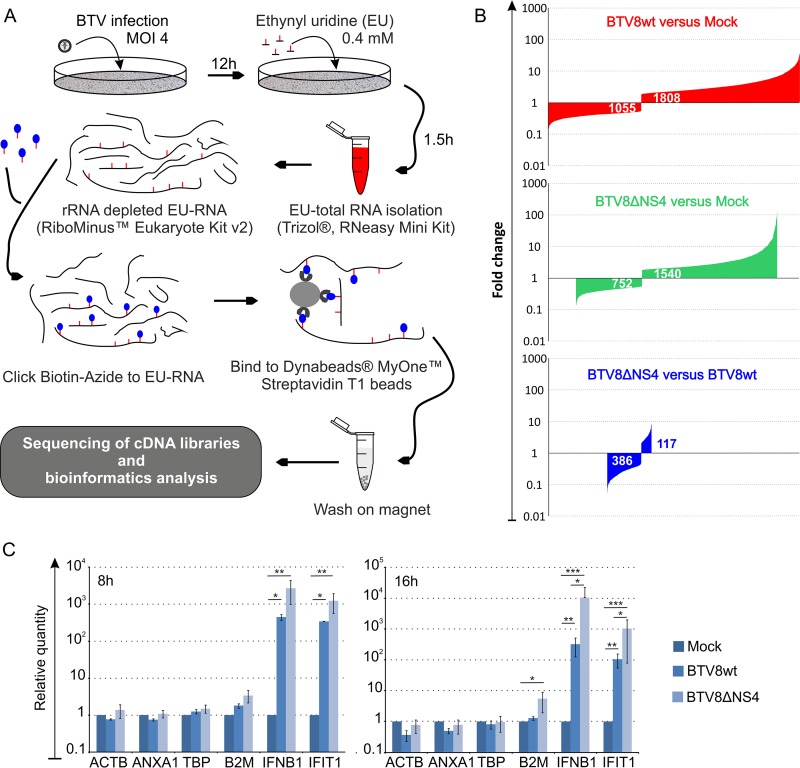
FIG 4.
RNA-seq of BTV8wt-, BTV8ΔNS4-, and mock-infected cells. (A) Schematic representation of RNA-sequencing workflow. A549 cells were mock infected or infected with BTV8wt or BTV8ΔNS4 at an MOI of 4. At 12 h p.i., nascent RNA was metabolically labeled with an analogue of uridine (EU) at 0.4 mM for 90 min. Total RNA was extracted and enriched by selectively depleting rRNA transcripts. The EU-labeled RNA was chemically linked to azide-modified biotin and then captured on streptavidin magnetic beads. Biotin-labeled RNA attached to the magnetic beads was used directly to construct a library and subsequently sequenced using the Ion Proton sequencer. Sequence reads were processed as described in Materials and Methods. (B) Plots representing DE genes according to their fold change values. Log2-fold changes of >1 were regarded as upregulated (q value [false-discovery rate] < 0.05), whereas changes of <1 were regarded as statistically downregulated (q value < 0.05). (C) Validation of RNA-seq by qRT-PCR. mRNA levels of ACTB, ANXA1, TBP, B2M, IFN-β (IFNB1), and IFIT1 were measured by qRT-PCR at 8 and 16 h p.i. as described in Materials and Methods. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way ANOVA; Tukey's multiple-comparison test). The error bars represent standard deviations.