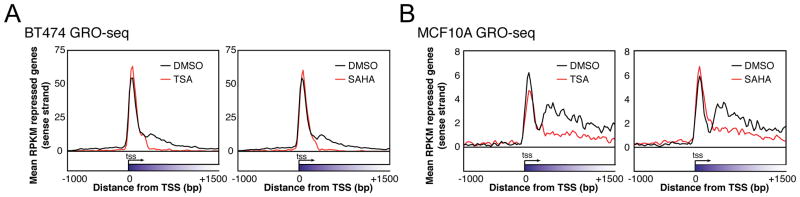
Figure 1. HDACIs block elongation of RNAP2 transcription in the genes they repress.
(A) Metagene plots of GRO-seq RPKM in the sense direction for repressed genes in BT474. Repressed genes had significantly (log-likelihood ratio P < 10−20, one GRO-seq experiment) reduced RPKM in the gene body (300 bp downstream of TSSs to gene ends). 6354 repressed genes were analyzed for TSA, and 7389 for SAHA.
(B) Same as (A) for MCF10A cells. There were 3866 repressed genes for TSA, and 4643 for SAHA.
See Figure S1 for GRO-seq statistics, RNAP2 ChIP-seq, GRO-seq of a shorter treatment of SAHA in BT474, SAHA blocking elongation in another cell line, and validation that SAHA and TSA repress transcription through their effects on HDACs.
See Table S1 for the number of genes common to each expression change subgroupings between the GRO-seq shown here and previously published GRO-seq data.
See Table S2 for median fold changes in subclassifications of gene expression changes for different factors by ChIP-seq in promoter regions.
See Table S3 for median fold changes in subclassifications of gene expression changes for different factors by ChIP-seq in gene body regions.