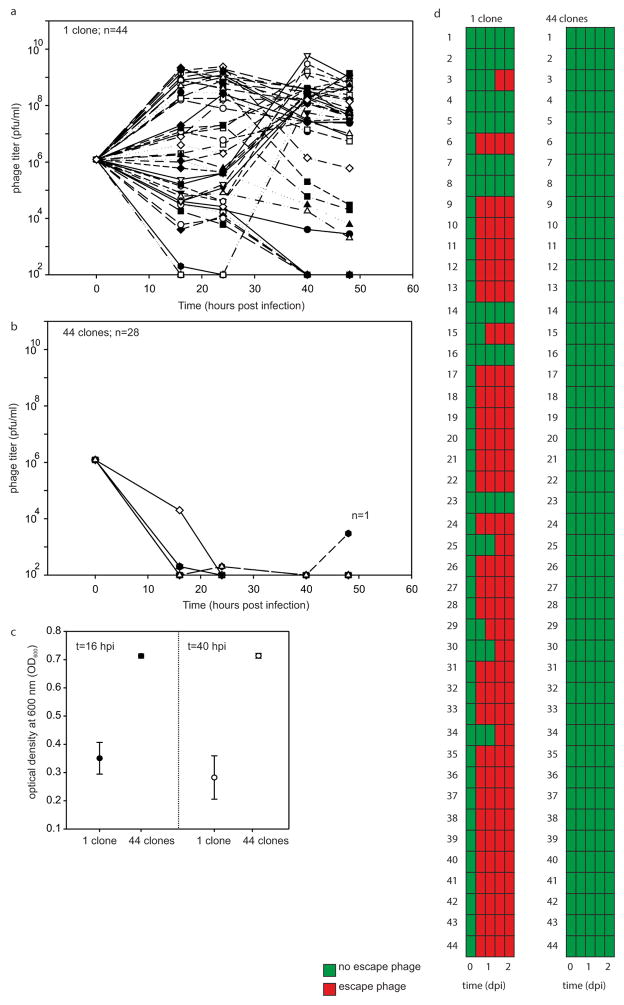
Extended Data Figure 2. Virus persistence inversely correlates with the level of CRISPR spacer diversity in CRISPR immune populations of S. thermophilus.
a, b, Virus titres (in plaque-forming units per millilitre) over time upon infection of a bacterial population consisting of a monoculture with CRISPR-mediated immunity (a; n =44 biological replicates) or 44-clone polycultures with CRISPR-mediated immunity (b; n = 28 biological replicates). Each clone is equally represented in each treatment. Each line indicates a biological replicate experiment. The limit of detection is 200 p.f.u. ml−1. c, Absorbance at 600 nm of monocultures and polycultures at 16 and 40 h.p.i. Error bars, 95% CI. d, Emergence of virus mutants that overcome CRISPR-mediated immunity after 0, 16, 24, 40 and 48 h.p.i. Green indicates no escape virus. Red indicates emergence of escape virus. Escape virus emerged in none of the 28 biological replicates of the polyculture experiments.