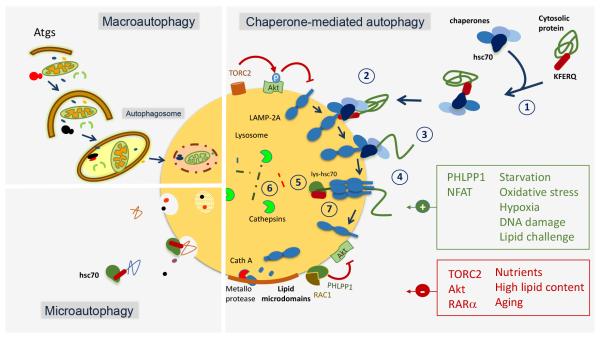
Figure 1. Schematic model of the different types of autophagy in mammals.
The three autophagic pathways that normally co-exist in most mammalian cells are depicted: Macroautophagy requires sequestration of cytosolic cargo by a limiting membrane that forms through conjugation of autophagy-related proteins (Atgs) and seals into a double membrane vesicle (autophagosome). Degradation occurs upon fusion of the autophagosome with the lysosome. In microautophagy, cytosolic cargo is trapped in small vesicles that form by invagination of the lysosomal membrane. A selective form of microautophagy is also depicted in which cytosolic proteins are selectively targeted by hsc70 to the microvesicles for internalization and degradation in the lumen. Details are shown of the individual steps that mediate degradation of cytosolic proteins via chaperone-mediated autophagy: 1) substrate recognition by hsc70; 2) binding to the cytosolic tail of LAMP-2A; 3) unfolding of the substrate protein by the chaperone complex; 4) multimerization of LAMP-2A to form the translocation complex; 5) substrate translocation with the assistance of the lysosome resident hsc70 (lys-hsc70); 6) substrate degradation and 7) disassembly of LAMP-2A from the translocation complex to initiate a new round of substrate binding and translocation. Regulation of CMA through the TORC2-Akt-PHLPP1 axis and through the degradation of LAMP-2A in lipid membrane microdomains through the combined action of a membrane metalloprotease and cathepsin A is shown. Activators and inhibitors of CMA are depicted in green and red, respectively. Abbreviations: Atg: autophagy-related proteins; Akt: protein kinase B; cath: cathepsin; hsc: heat shock cognate protein; LAMP: lysosome-associated membrane protein; NFAT: nuclear factor of activated T cells; PHLPP1: PH domain and leucine rich repeat protein phosphatase 1; RAC1: ras related C3 botulinum toxin substrate1; RARα: retinoic acid receptor alpha; TORC: target of rapamycin complex.