Fig. 2.
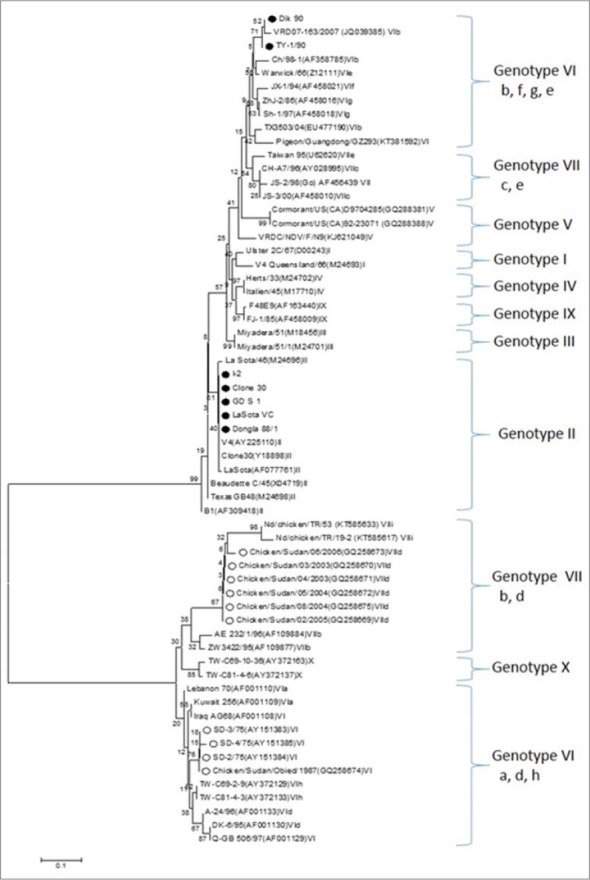
Rooted phylogenetic tree of the nucleotide sequences of the fusion protein gene of Newcastle disease virus strains. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with the sum of branch length = 2.20497769 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. The analysis involved 61 nucleotide sequences. Evolutionary analyses were conducted in MEGA7. Isolates analyzed in this study are marked with filled circle, whereas those reported earlier from the Sudan are marked with empty circles.
