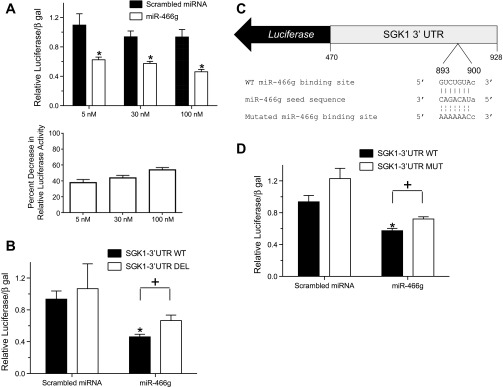
Fig. 4.
miR-466g interacts with its target sequence in the 3′-UTR of SGK1. A: HEK-293 cells were cotransfected with a luciferase reporter containing the 3′-UTR of SGK1 (SGK1-3′-UTR WT) and increasing doses of a scrambled miRNA or miR-466g. B: HEK-293 cells were cotransfected with SGK1 3′-UTR WT or a luciferase reporter in which the putative miR-466g binding site in the SGK1 3′-UTR was deleted (SGK1-3′-UTR DEL) and either scrambled miRNA or miR-466g (10−7 M). C: schematic of a luciferase reporter in which serial point mutations have been introduced into the putative miR-466g seed binding site (nucleotides 893–900) of the 3′-UTR of SGK1 (SGK1-3′-UTR MUT). D: HEK-293 cells were cotransfected with SGK1-3′-UTR WT or SGK1-3′-UTR MUT and either scrambled miRNA or miR-466g (3.0 × 10−8 M). In all experiments, luciferase activity was first normalized to β-galactosidase activity and whole cell protein concentration and then normalized to background luciferase/β-galactosidase activity in cells transfected with luciferase vector. Values are means ± SE, from at least 5 independent experiments. *P < 0.05 between cells transfected with scrambled miRNA and miR-466g. +P < 0.05 between cells transfected with SGK1-3′-UTR and either SGK1-3′-UTR DEL or SGK1-3′-UTR MUT.