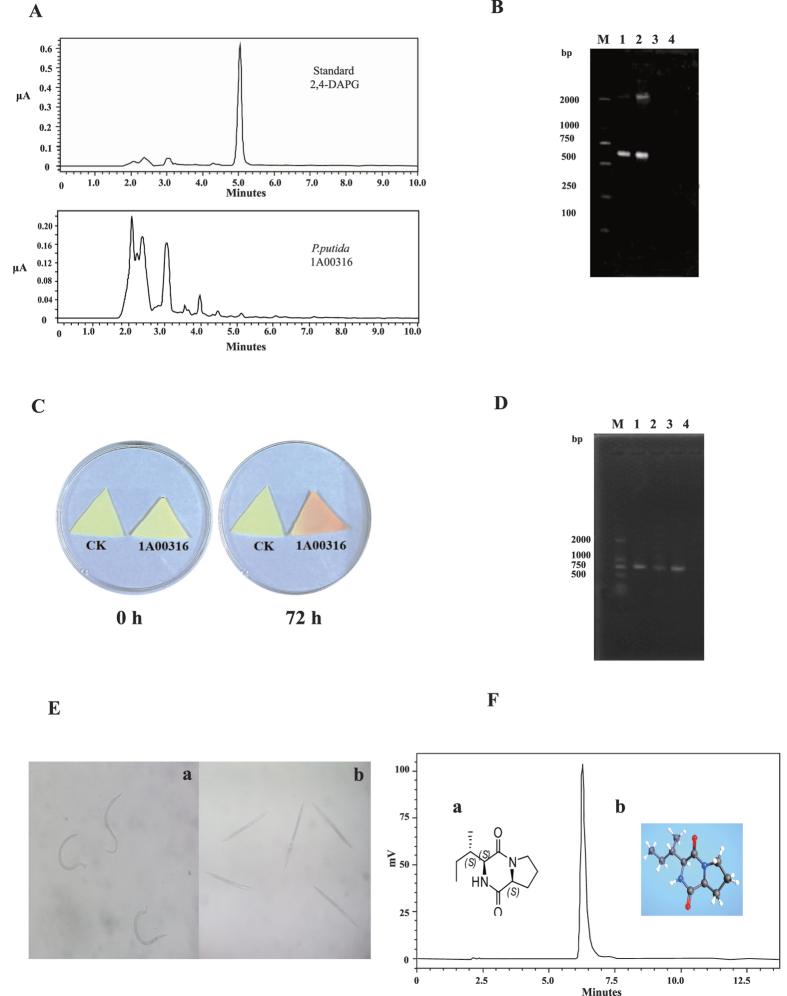
Figure 4. Experimental analysis of small molecule of nematicidal factors in P. putida 1A00316.
(A) HPLC chromatogram of standard 2,4-DAPG and crude extracts of P. putida 1A00316 fermentation broth. (B) PCR amplification of the phlD gene fragment. M represents DL 2000, lines 1–3 represent the DNA of P. fluorescens CHA0, P. fluorescens Pf-5, and P. putida 1A00316, respectively, and line 4 is the water control. (C) HCN detection with biochemical methods. HCN produced by P. putida 1A00316 in microtiter plates with indicator paper at 0 and 72 h. Yellow paper indicated that P. putida 1A00316 could not produce HCN, while orange indicated that it could produce HCN. (D) PCR amplification of the hcnB gene fragment. M represents the marker of DL 2000, lines 1–3 represent the DNA of P. fluorescens CHA0, P. fluorescens Pf-5, and P. putida 1A00316, respectively, and line 4 is the water control. (E) The light microscopy results for M. incognita treated with the small metabolite cyclo-(l-Ile-l-Pro) produced by P. putida 1A00316. ‘a’ and ‘b’ treated by cyclo-(l-Ile-l-Pro) for 0 h and 24 h respectively. (F) Structures of cyclo-(l-Ile-l-Pro) and HPLC results for purified cyclo-(l-Ile-l-Pro) from P. putida 1A00316. ‘a’ and ‘b’ represent the two-dimensional and three-dimensional structures of cyclo-(l-Ile-l-Pro), respectively.