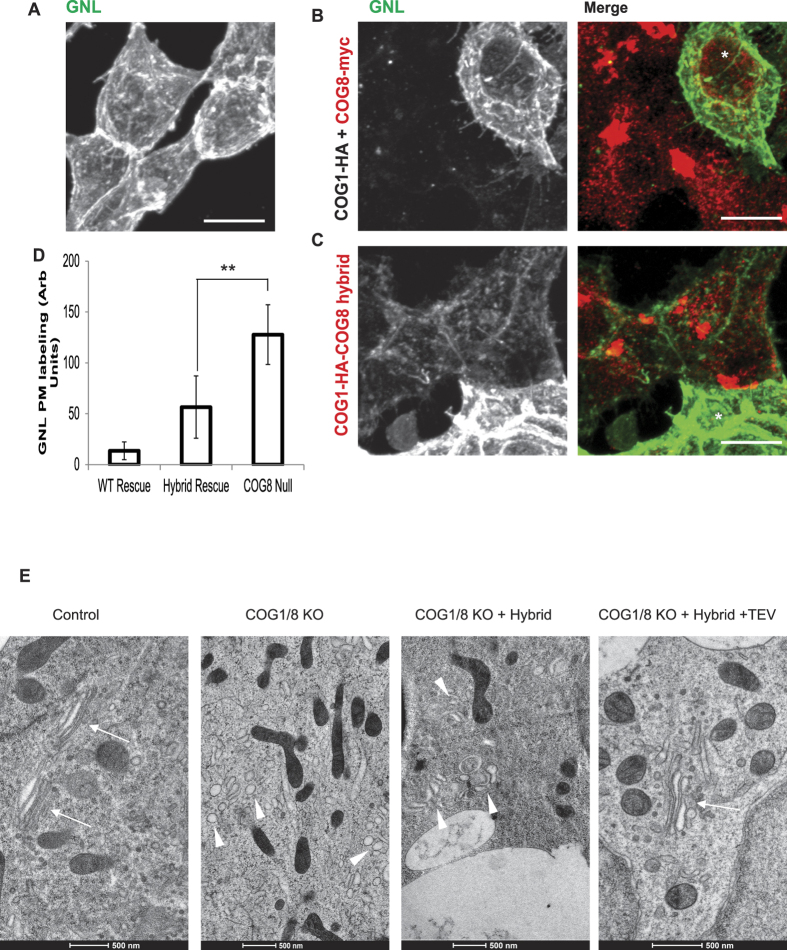
Figure 8. COG complex disassembly is critical for its role in Golgi glycosylation and stability.
ΔCOG8 cells (A) were transiently transfected with siRNA against COG1 (B,C) and then co-transfected with plasmids encoding COG1-3Myc and COG8-3Myc (B) or COG1-HA-GS-COG8 hybrid (C). 72 hours after transfection cells were fixed, surface labeled with GNL-647, and stained with anti-Myc antibodies for confocal microscopy analysis. The recovery of the ΔCOG mediated glycosylation defect was quantified by line scan analysis using Fiji plot profile and the averages are shown in the graph (D). 12–14 cells were analyzed with three measurements per cell. Error bars denote standard deviation. * Denotes control un-transfected cell. (E) TEM images of the Golgi region. Control HEK293T cells (control), ΔCOG1/ΔCOG8 (COG1/8 KO) or ΔCOG1/ΔCOG8 cells transfected with the COG1-HA-GS-COG8 hybrid alone (COG1/8 KO + hybrid), or transfected with COG1-HA-GS COG8 Hybrid and hTEV plasmid (COG1/8 KO + Hybrid + TEV) were grown on sapphire discs for 3 days, subjected to the HPF-FS procedure and embedded in resin. Thin-sections were cut at 50 nm and images at 11,500x. Golgi stacks we observed in 80% of control cells (n = 10) and in 37% of ΔCOG1/Δ COG8 + Hybrid + TEV cells (n = 8) but not in ΔCOG1/Δ COG8 or ΔCOG1/Δ COG8 + hybrid cells (n = 10). Note Golgi stacks (arrows) and Golgi-like dilated membranes (arrowheads).