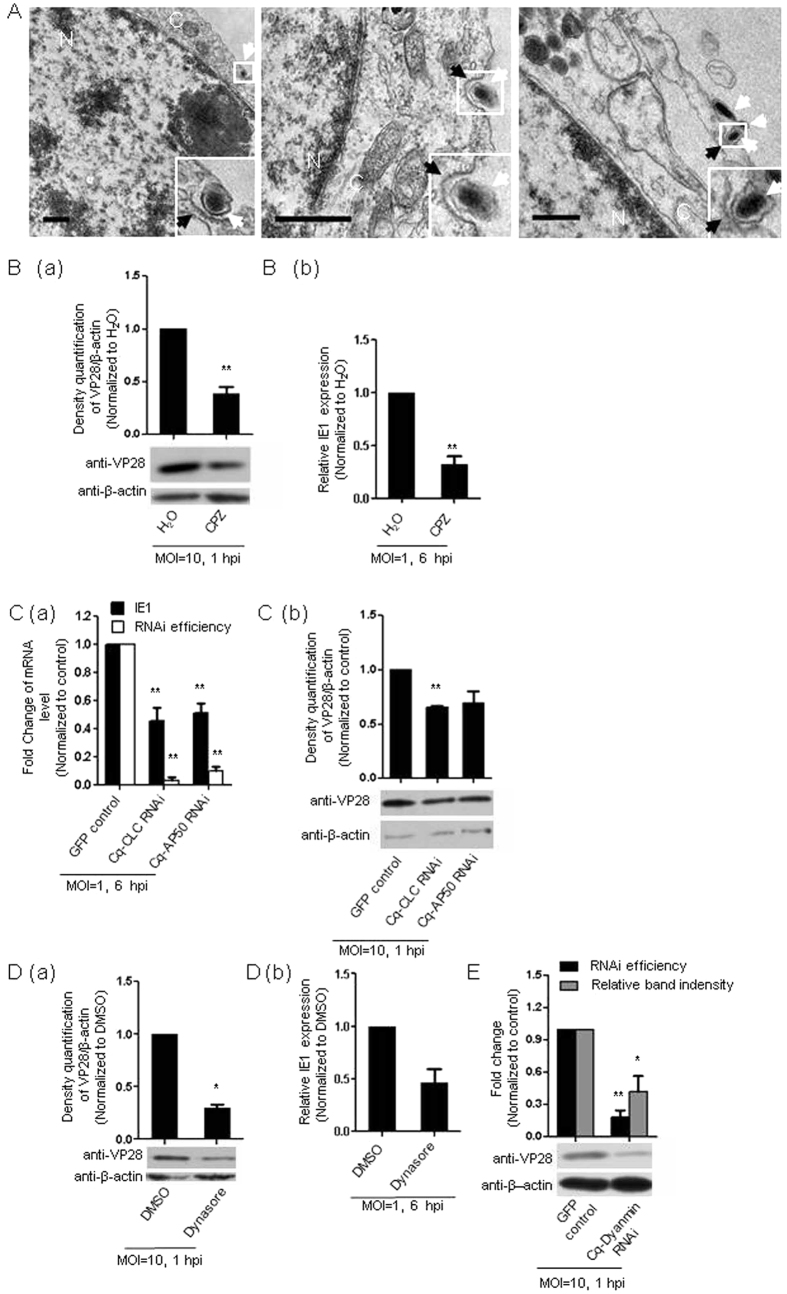
Figure 1. WSSV entry into Hpt cell via CME.
(A) WSSV virions captured in cell invaginations resembling clathrin-coated pits at 0.5 hpi by TEM. Black arrows indicate clathrin-coated pits, and white arrows indicate WSSV virions. The bars indicate 0.5 μm. C: cytoplasm; N: nucleus. (B) Inhibition of WSSV entry by pretreatment of the cells with CPZ. (a) The major viral envelope protein VP28 was immunoblotted with a monoclonal antibody against VP28 at 1 hpi (lower panel). (b) Suppression of WSSV replication by pretreating the cells with CPZ. Suppression was evaluated by assessing relative IE1 gene expression at 6 hpi using qRT-PCR. (C) (a) Inhibition of WSSV replication via knockdown of Cq-CLC or Cq-AP50 as assessed by qRT-PCR at 6 hpi to quantify the relative IE1 gene expression. The RNAi efficiency of Cq-CLC and Cq-AP50 was also examined by qRT-PCR. (b) Inhibition of WSSV entry via knockdown of Cq-CLC or Cq-AP50 as determined by immunoblotting against the viral envelope protein VP28 at 1 hpi (lower panel). (D) (a) Suppression of WSSV entry by pretreating the cells with dynasore determined by immunoblotting against the viral envelope protein VP28 at 1 hpi (lower panel). (b) Inhibition of WSSV replication by pretreating the cells with dynasore to assess the relative IE1 gene expression by qRT-PCR at 6 hpi. E: RNAi efficiency of Cq-dynamin examined by qRT-PCR (dark bars in upper panel). The suppression of WSSV entry via gene knockdown of Cq-dynamin was determined from the relative quantification of the viral envelope protein VP28 at 1 hpi by immunoblotting (lower panel). The band intensities of three independent experiments were calculated using the Quantity One program (upper panel in B(a), C(b), D(a) and gray bars in upper panel in E). All the results were observed in at least three independent experiments, and one of the representative results is shown. The data are presented as the mean ± SEM from at least three independent experiments and were analyzed by Student’s t test (*P < 0.05, **P < 0.01).