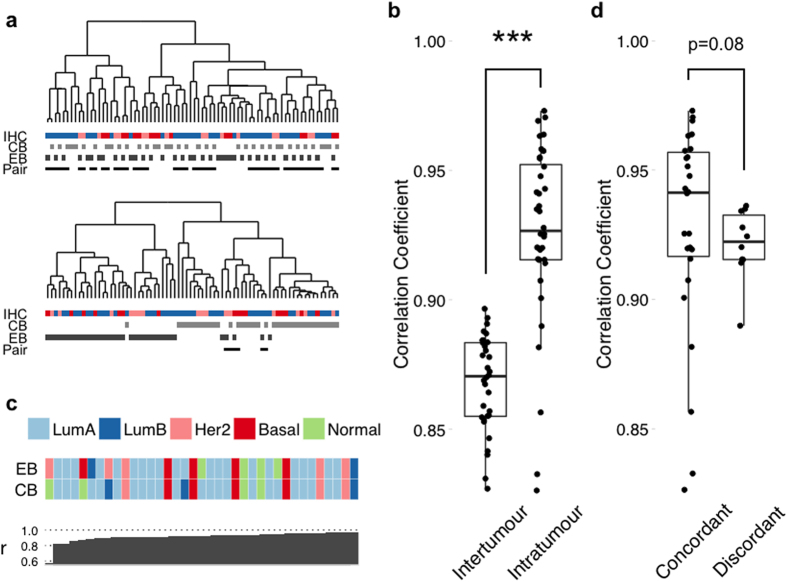
Figure 1. Evidence of treatment independent variation between breast cancer diagnostic core biopsies and surgical excision samples.
(a) Hierarchical clustering of the 37 patient-matched diagnostic core and excision biopsy samples using the 500 most variable genes (upper) and a SAM derived signature of 50 genes consistently differentially expressed between core and excision biopsies (lower). Bars represent IHC status (ER+/Her2− = Blue; ER+/Her2+ = Pink; ER-/Her2− = Red) or biopsy method. Lower-most bar indicates where sample pairs have co-aggregated. Two-thirds (25/37) of the pairs cluster at the first level of the upper dendrogram, whereas pairwise association is lost in 31/37 cases for the lower. (b) There is a significantly stronger correlation between biopsy pairs (intra-tumour) than between different tumours (mean inter-tumour). ***p < 0.001. (c) Discordance in molecular subtype assignment between core and excision biopsies. Patients are ranked left to right by pairwise correlation. Colours represent SSP subtypes (Luminal A = Dark blue; Luminal B = Light Blue; Her2 = Pink; Basal = Red; Normal = Green). (d) Sample pairs were called discordant when Biopsy A ≠Biopsy B for at least 4/5 classifiers. Comparison of concordant vs. discordant pairwise correlations then revealed an inverse relationship between correlation and discordancy.