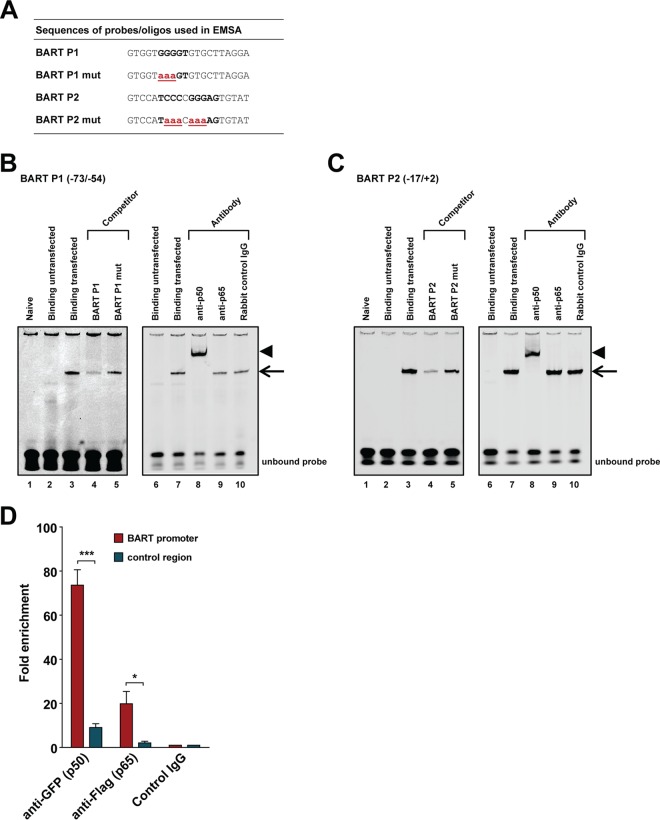
FIG 2.
NF-κB binds to NF-κB elements in the BART promoters. (A) Nucleotide sequences of the double-stranded probes and oligonucleotides used in the competition experiments shown below. The NF-κB binding sites in the BART P1 and P2 promoters, consisting of NF-κB half-site sequence(s), are shown in bold uppercase letters, and the mutated nucleotides are in red bold lowercase letters and underlined. (B and C) IRDye 700-labeled double-stranded synthetic oligonucleotides corresponding to the BART P1 region from position −73 to −54 or the BART P2 region from position −17 to +2, as indicated, of the B95-8 sequence were incubated with nuclear extract from HEK 293T cells transfected with plasmids expressing EGFP-tagged p50 and Flag-tagged p65 and subjected to EMSA. The black arrow indicates the NF-κB/probe complex on the gel. Lanes 2 and 6 show the binding pattern of nuclear extract from untransfected cells, while lanes 3 and 7 show the binding pattern of nuclear extract from p50/p65 transfected cells. Lanes 4 and 5 show the binding of the probe when an excess (300-fold) of unlabeled wild-type or mutant competitor oligonucleotide was added to the binding reaction mixture. Supershift experiments with antibodies were performed as indicated above the gels, with supershifted complexes indicated by an arrowhead in lanes 8 of panels B and C. (D) ChIP assay of NF-κB p50 and p65 with the BART promoter. HEK 293T cells were transfected with a plasmid containing the BART P1 promoter sequence and EGFP-tagged p50 and Flag-tagged p65. Results of real-time PCR analysis and ChIP assays with antibodies specific for GFP or Flag or rabbit control IgG are shown. The results are expressed as fold enrichment, where the value for the rabbit control IgG was set to 1. A genomic region ∼5 kb upstream of the IL-8 promoter was used as a negative control. The averages and standard errors of the means from three independent experiments are shown, with all samples being analyzed in triplicate. P values were obtained from a two-tailed Student's t test (*, P < 0.05; ***, P < 0.001).