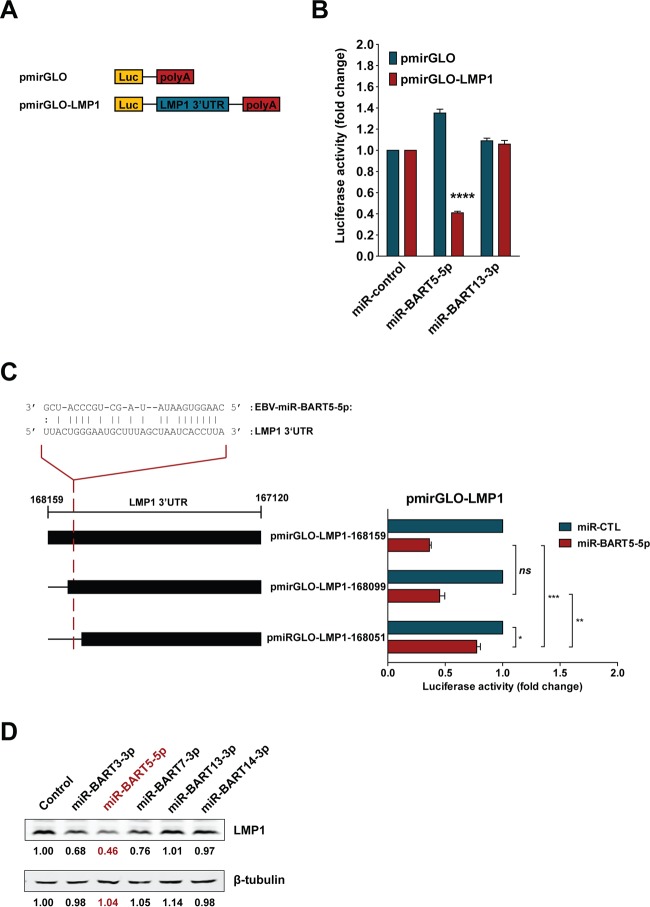
FIG 7.
EBV-encoded miR-BART5-5p modulates LMP1 expression. (A) The pmirGLO reporter plasmid constructs used to assess the effect of miR-BART5-5p on the LMP1 3′ UTR. (B) HEK 293T cells were transfected with pmirGLO-LMP1 or empty pmirGLO reporter, together with 50 nM total synthetic miRNA (miR-control), miR-BART5-5p, or miR-BART13-3p duplexes. Reporter activity was determined 24 h after transfection. Firefly/Renilla ratios were normalized against the same reporter transfected with the negative-control miRNA, and results are expressed as fold change in luciferase activity. (C) Sequence alignment of miR-BART5-5p and the predicted binding site on the LMP1 3′ UTR (35). HEK 293T cells were cotransfected with a pmirGLO-LMP1 or pmirGLO-truncated LMP1 3′ UTR reporter and either 50 nM miR-CTL or miR-BART5-5p. Firefly/Renilla ratios were normalized to levels of the same reporter transfected with miR-CTL and expressed as fold change in luciferase activity. The results are the averages and standard errors of the means from three independent experiments. P values are from a two-tailed Student's t test comparison of results with the BART miRNA to those with the control miRNA (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant). (D) HEK 293T cells stably expressing LMP1, including its 3′ UTR, were transfected with 50 nM synthetic miRNA (control), miR-BART3-3p, miR-BART5-5p, miR-BART7-3p, miR-BART13-3p, or miR-BART14-3p duplexes. Expression of LMP1 was detected by immunoblotting using specific antibodies, and the intensities of bands were determined using Image Studio Lite software. Levels of β-tubulin were determined as a loading control. The intensities of the bands were normalized to the level of the negative-control miRNA and are expressed as fold change in protein expression.