Summary
Inhibition of sulfotransferases by pentachlorophenol, and siRNA knockdown of the SULT1A1 gene in human kidney (HK-2) and human fibroblast (GM00637) cell lines reduce significantly the levels of aristolactam-DNA adducts and toxicities induced by aristolochic acid I and its metabolite, N-hydroxyaristolactam I.
Abstract
Aristolochic acids (AA) are implicated in the development of chronic renal disease and upper urinary tract carcinoma in humans. Using in vitro approaches, we demonstrated that N-hydroxyaristolactams, metabolites derived from partial nitroreduction of AA, require sulfotransferase (SULT)-catalyzed conjugation with a sulfonyl group to form aristolactam-DNA adducts. Following up on this observation, bioactivation of AA-I and N-hydroxyaristolactam I (AL-I-NOH) was studied in human kidney (HK-2) and skin fibroblast (GM00637) cell lines. Pentachlorophenol, a known SULT inhibitor, significantly reduced cell death and aristolactam-DNA adduct levels in HK-2 cells following exposure to AA-I and AL-I-NOH, suggesting a role for Phase II metabolism in AA activation. A gene knockdown, siRNA approach was employed to establish the involvement of selected SULTs and nitroreductases in AA-I bioactivation. Silencing of SULT1A1 and PAPSS2 led to a significant decrease in aristolactam-DNA levels in both cell lines following exposure to AA-I, indicating the critical role for sulfonation in the activation of AA-I in vivo. Since HK-2 cells proved relatively resistant to knockdown with siRNAs, gene silencing of xanthine oxidoreductase, cytochrome P450 oxidoreductase and NADPH:quinone oxidoreductase was conducted in GM00637 cells, showing a significant increase, decrease and no effect on aristolactam-DNA levels, respectively. In GM00637 cells exposed to AL-I-NOH, suppressing the SULT pathway led to a significant decrease in aristolactam-DNA formation, mirroring data obtained for AA-I. We conclude from these studies that SULT1A1 is involved in the bioactivation of AA-I through the sulfonation of AL-I-NOH, contributing significantly to the toxicities of AA observed in vivo.
Introduction
Aristolochic acids (AA) became widely recognized as human nephrotoxins in the early 1990s when ~100 Belgian women developed kidney disease following ingestion of herbal remedies as part of a slimming regimen (1). Subsequently, ~50% of these individuals developed upper urinary tract urothelial carcinomas (UTUC) (2). The unique pathophysiology of this disease, initially named Chinese herbs nephropathy, provided important clues that later were used to solve the long-standing mystery of an environmental disease, so-called Balkan endemic nephropathy (3–5). These apparently unrelated syndromes had in common exposure to AA, a product of Aristolochia plants. In Belgium, AA was ingested as a component of a Chinese traditional medicine and, in the Balkans, as a contaminant of wheat flour used for baking homemade bread (1,4,6). Both disorders are now referred to as aristolochic acid nephropathy (AAN) (7,8).
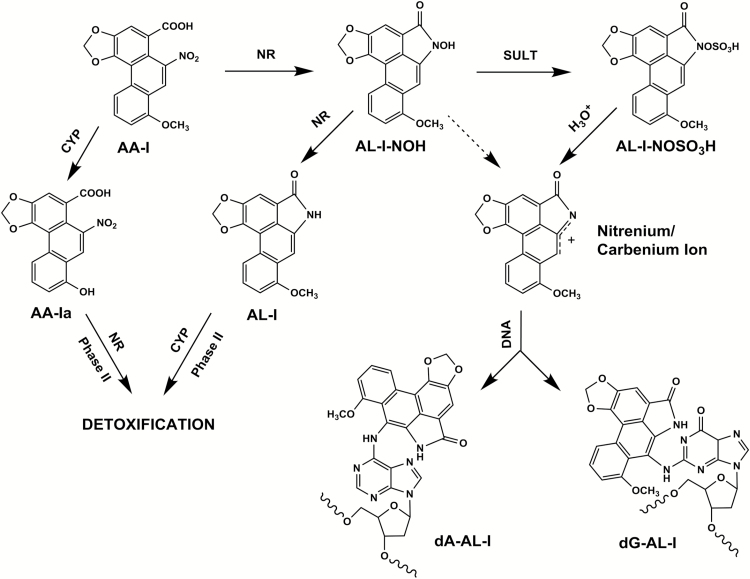
AA-I (Figure 1) and its 8-demethoxylated analog, AA-II, are nitrophenanthrene carboxylic acids produced exclusively by plants of the Aristolochiaceae family (9,10). The term AA is used to designate a mixture of these structural analogs. Although both compounds are carcinogenic in rodents (11–13) and mutagenic in bacterial (14) and mammalian cells (15), only AA-I displays nephrotoxic properties in rodents (16,17).
Figure 1.
Pathways for AA-I bioactivation and detoxification. AA-I undergoes four-electron NR to form AL-I-NOH, followed by -O-sulfonation catalyzed by SULTs. Sulfonyloxyaristolactam (AL-I-N-OSO3H) solvolizes readily in aqueous solution, affording the cyclic nitrenium/carbenium ion that, in turn, reacts with DNA to form dA-AL-I and dG-AL-I adducts. AL-I-NOH shows low reactivity with DNA (indicated with a dashed line), which may account for the incorporation of AA-I into DNA in the absence of SULTs. Six-electron NR produces aristolactam (AL-I), which itself undergoes oxidative demethylation and/or Phase II reactions yielding to easily excretable conjugates. In parallel, CYP1A2/1 can cleave the 7-O-methoxy group, creating the non-reactive AA-Ia, a substrate for NR and/or Phase II detoxification reactions.
The following aristolactam (AL)-DNA adducts have been detected in cells and tissues of rodents exposed to AA: 7-(deoxyadenosin-N6-yl)aristolactam I (dA-AL-I), 7-(deoxyguanosin-N2-yl)aristolactam I (dG-AL-I) (Figure 1), 7-(deoxyguanosin-N2-yl)aristolactam II (dG-AL-II) and 7-(deoxyadenosin-N6-yl)aristolactam II (dA-AL-II) (18,19). dA-AL-II directs incorporation of dAMP opposite the adduct, leading to A to T transversion mutations (20). The dA-AL:dT pair is highly resistant to global genomic nucleotide excision repair (21), generating useful biomarkers of AA exposure and effect, namely, highly persistent dA-AL-I adducts in renal cortex tissues and A:T to T:A transversions in the non-transcribed strand of the TP53 gene in tumor cells. These biomarkers, established in our studies of Balkan endemic nephropathy (4,5), were used to implicate AA in the high incidence of UTUC cases reported in Taiwan (22). Subsequently, the signature A to T mutation was shown to occur genome wide in tumor DNA obtained from UTUC patients in Taiwan (23,24). These studies revealed also that the mutational load exerted by AA exposure is much higher than that linked to other Group I carcinogens, such as tobacco smoke and ultraviolet light (25). Recently, the AA-signature mutation was found in hepatocellular (24) and renal cell carcinomas (26); thus, the role of AA in tumorigenesis in non-urothelial tissues is strongly implied.
Since only 5–10% of individuals exposed to AA are prone to developing AAN/UTUC (27), and genes responsible for the metabolism of xenobiotics may confer susceptibility to such compounds, it was important to elucidate fully the pathways by which AA-I is biotransformed. There are two major routes for AA-I metabolism, oxidation and reduction (Figure 1). The former predominates in hepatic tissues, involving oxidative demethylation of AA-I by CYP1A2/1, leading to formation of the non-toxic 8-OH-AA-II (AA-Ia) that, in turn, serves as a substrate for nitroreduction (NR) and/or conjugation with glucuronic and sulfuric acids, forming soluble, excretable metabolites (28–32).
NR of AA-I produces inactive and active metabolites of AA-I. Inactive intermediates include aristolactam I (AL-I) (Figure 1) and 8-hydroxyaristolactam II, end products of AA-I NR and demethylation (32). Their glucuronides have been detected in feces and urine of various mammalian species exposed to AA (30,31). As postulated for other nitroaromatic compounds, partial NR of AA-I forms the hydroxylamine [N-hydroxyaristolactam I (AL-I-NOH)] that, until recently, was considered the immediate precursor of the electrophilic cyclic nitrenium ion in the formation of AL-I-DNA adducts (Figure 1) (33–35). The following oxidoreductases have been shown to metabolize AA-I: NAD(P)H:quinone oxidoreductase 1 (NQO1), xanthine oxidase (XDH), prostaglandin H synthase, NADPH:CYP reductase (POR) and CYP1A1/2 (36). Nevertheless, evidence for their direct involvement in AA-I bioactivation in vivo is thus far lacking or controversial (37,38).
Hydroxylamine metabolites of nitroarenes acquire increased reactivity upon sulfonation (39,40). Variable individual sensitivity to the toxic effects of AA among human populations suggests the role of yet unknown genetic variants. In this regard, the potential involvement of sulfotransferases (SULTs) in AA bioactivation is of considerable interest. Despite the inherent plausibility of the Phase II activation pathway (41), the Stiborova’s laboratory reached an opposite conclusion (42) regarding the role of SULTs in AA mutagenicity and reactivity. We attempted to resolve this discrepancy by demonstrating in vitro that N-hydroxyaristolactams require sulfonation by murine and human SULTs, forming aristolactam-DNA adduct with high efficiency (Figure 1) (43). In this study, we extend this important observation to human cells.
Here, using a SULT inhibitor and also a gene knockdown approach, we examine the involvement of SULTs in the bioactivation of AA-I and AL-I-NOH in human kidney proximal tubules (HK-2) and human fibroblast (GM00637) cell lines. Additionally, the effect of silencing of selected nitroreductases was explored in GM00637 cells. Our experiments provide strong evidence for the critical role of SULTs, in particular SULT1A1, in the formation of AL-I-DNA adducts and cytotoxicity of AA-I and AL-I-NOH in human cells.
Materials and methods
Caution: γ-32P-ATP, AA-I, AL-I-NOH and pentachlorophenol (PCP) should be handled with appropriate safety measures.
Chemicals and enzymes
AA-I, AL-I-NOH, and dG-AL-II- and dA-AL-II-containing oligonucleotides were synthesized in our laboratory as described earlier (20,44). γ-32P-ATP (6000 Ci/mmol) was supplied by PerkinElmer (Boston, MA). Phosphodiesterase II and P1 nuclease used for DNA digestions were obtained from MP Biomedicals (Santa Ana, CA); micrococcal nuclease and potato apyrase were obtained from Sigma–Aldrich (St Louis, MO). 3′-Phosphatase mutant polynucleotide kinase was purchased from New England Biolabs (Ipswich, MA).
ATP luminescence kit, PCP, phosphate-buffered saline and dimethyl sulfoxide (DMSO) were purchased from Sigma–Aldrich. All reagents were of ACS or molecular biology grades.
Cell culture
Authentic human kidney proximal tubule (HK-2) and human skin fibroblast (GM00637) cell lines, obtained from Coriell Cell and ATCC repositories, respectively, were maintained in Dulbecco’s modified Eagle’s medium (DMEM/F-12, HyClone, Thermo Scientific), supplemented with fetal bovine serum (10%, vol/vol), under standard conditions, 5% (vol/vol) CO2 at 37°C. Medium without serum was used for exposure to AA-I and AL-I-NOH as described below.
siRNA source and quantitative PCR
ON-TARGETplus SMARTpool siRNAs used for silencing expression of human SULT1A1, SULT1A2, SULT1C2, SULT1C4, SULT1B1, PAPSS2, NQO1, POR, XDH genes and non-targeting (NT) siRNA (Supplementary Table S1, available at Carcinogenesis online) were purchased from Dharmacon GE Healthcare (Lafayette, CO).
Total RNA from cells was isolated by RNeasy mini kit (Qiagen). Complementary DNA was synthesized by QuantiTect reverse transcription kit (Qiagen), using random primers. QuantiTect SYBR green PCR kit (Qiagen) was used for quantitative PCR (qPCR) conducted on MJ Research DNA Engine Opticon 2 machine. PCR conditions were as follows: 15min at 95°C, followed by 45 cycles of 15s at 94°C, 30s at 60°C and 30s at 72°C. The size of the expected product was verified by agarose gel electrophoresis. DNA primers for GAPDH and PAPSS2 amplification were obtained from Origene Technologies (Rockville, MD). Other primers were custom designed and synthesized by Eurofins Genomics. For oligonucleotide pairs, see Supplementary Table S1, available at Carcinogenesis online.
To estimate the efficiency of siRNA-mediated gene silencing, complementary DNA from cells treated with NT siRNA was serially diluted and threshold cycles values (C t) for GAPDH, used as an internal control, and for a gene of interest were obtained by qPCR. C t values for GAPDH and a gene of interest were obtained using complementary DNA prepared from cells treated with gene-specific siRNA. Calibration curves were constructed to estimate the relative amounts of GAPDH and genes of interest in target cells. The relative amounts of the gene of interest before and after knockdown were normalized to corresponding values for GAPDH. The fold-change in expression was derived by dividing normalized values in the silenced sample by those of the NT control (Supplementary Table S2, available at Carcinogenesis online).
siRNA transfections and AA exposure
Prior to the experiment, GM00637 cells (3×106), hereafter referred to as GM637, were seeded in a 75cm2 flask, cultured overnight and transfected by the Lipofectamine RNAiMAX reagent (Life Technologies) with 600 pmol of one of the following siRNAs: NT, PAPSS2, SULT1A1, SULT1A2, SULT1A2 and SULT1A1 (double knockdown), SULT1B1, NQO1, POR or XDH. After 72h, media was removed. Cells were washed with serum-free DMEM/F-12 media, then exposed to 25 µM of AA-I or AL-I-NOH for 24h under standard conditions. Each experiment included one NT and two or three gene-specific siRNAs and was conducted independently with three or more cell passages. Following exposure, cells from each flask were collected by trypsinization and divided into the following groups: two for DNA isolation and adduct analysis and one for RNA isolation and qPCR. DNA was isolated by DNeasy Blood and Tissues kit purchased from Qiagen (Valencia, CA) and analyzed for the presence of AL-I-DNA adducts by 32P-postlabeling, as described below.
HK-2 cells were treated similarly to GM637 with minor adjustments. HK-2 cultures were exposed to one of the following siRNAs for 48 h: NT, PAPSS2, SULT1A1, SULT1C2, SULT1C4, SULT1B1 and NQO1. Due to relative resistance to siRNA knockdown, experiments with HK-2 were limited to AA-I administration (6h) and did not include SULT1A2, POR and XDH silencing.
32P-postlabeling polyacrylamide gel electrophoresis adduct analysis
DNA adduct levels were determined as described previously (19,43) with minor modifications. DNA (5 µg) was digested in a solution (100 µl) composed of 20mM sodium succinate buffer (pH 6.0), 8mM CaCl2, spleen phosphodiesterase II (0.015 units) and micrococcal nuclease (2 units). Samples were incubated for 6h at 37°C, followed by addition of 1 µl of 100mM ZnCl2 and 1 unit of nuclease P1, and incubation for 1h at 37°C. Digested DNA was enriched for adducts by butanol extraction. Butanol fractions were evaporated to dryness, then dissolved in water. AL-I-DNA adducts were postlabeled with γ-32P-ATP by 3′-phosphatase mutant polynucleotide kinase, then loaded on 30% nondenaturating acrylamide gels. Results were visualized by phosphorimaging. To quantify AL-I-DNA adduct levels, standard oligonucleotides containing a single dG-AL-II or dA-AL-II adduct were digested in parallel with the DNA samples (21,43). As a control, DNA samples from previous experiment were included in a subsequent analysis and digested in parallel with DNA prepared from newly exposed cells.
Assays for a genotoxicity of AA-I and AL-I-NOH in HK-2 cells
HK-2 cultures were grown to confluence on 60mm plates. Prior to the experiment, cells were washed thoroughly in media without serum and exposed to 5, 12.5 or 25 µM of AA-I or AL-I-NOH for 4, 8, 24, 32 and 48h under standard conditions. To estimate the effect of PCP presence on AL-I-DNA formation in HK-2 cells, cell cultures were exposed for 6h to 12.5 µM of AA-I or AL-I-NOH, with or without PCP, 0.4–25000nM. Following exposure, cells were washed with phosphate-buffered saline, collected and stored at −80°C until used for DNA isolation and adduct analysis as described above.
To determine the dependence of rates of AL-I-DNA formation on PCP presence, cells were treated with 12.5 and 25 µM of AA-I or AL-I-NOH for 4, 8, 24 and 48h. Exposures were conducted with or without PCP, 250nM for AA-I and 1000nM for AL-I-NOH. Isolated DNA was analyzed for the presence of adducts.
All experiments were conducted independently on three different cell passages. Every passage included three plates for each exposure condition.
Assay for cytotoxicity of AA-I and AL-I-NOH in HK-2 cells
HK-2 cells were grown to confluence on 24-well plates. Prior to the experiment, cells were washed in media without serum, then treated with 1–50 µM of AA-I or AL-I-NOH, with or without addition of 250 or 1000nM PCP, respectively. Each exposure was conducted for 24 and 48h under standard conditions. Cells treated with plain media, plain media with 0.1% DMSO and plain media with 250 or 1000nM PCP were used as mock controls. In addition, the effect of varying doses of PCP (250–25000nM) on ATP content, measured 24 and 48h after exposure, was explored. Cell viability assays were performed as described previously (21). Briefly, following exposure, cells were washed with PBS, lysed and ATP levels were evaluated by luminescence. Cytotoxicity was defined as the ratio of ATP levels in cells treated with a test compound to ATP levels in the DMSO control. When PCP was coadministered with AA-I or AL-I-NOH, cells treated with the corresponding concentration of PCP were used as the reference control. Experiments were carried out on at least two different cell passages. Each passage included three wells per each exposure.
Data analysis
Sigma Plot v8.0 (SPSS) was used to plot results. Linear regression was employed to estimate the rates of adduct formation and toxicity in HK-2 cells exposed to AA-I and AL-I-NOH. To obtain IC50 and EC50 values, a four-parameter logistic regression analysis was used. Results are reported as mean values with SD. The two-tailed t-test for independent samples was used to compare knockdown conditions to NT control. Conditions with P values <0.05 were considered to be significantly different from controls.
Results
Cytotoxicity and genotoxicity of AA-I and AL-I-NOH in HK-2 cells
HK-2 cells are sensitive to AA-I. Microarray analysis shows that this cell line expresses mRNA from genes coding for various SULT isoforms and one of the two genes responsible for 3′-phosphoadenosine-5′-phosphosulfate synthesis, PAPSS2(K. Dickman, unpublished data). For AL-I-NOH, NR is no longer required and toxicities are expected to be similar to or greater than AA-I. Recently, we reported that a decline in ATP levels in cultured cells exposed to AA-II reflects its cytotoxicity, whereas AL-II-DNA adducts are good biomarkers for genotoxicity (21).
HK-2 cells, grown on 24-well plates, were treated with AA-I or AL-I-NOH (0–50 µM), and the ATP content for each well was evaluated following 24–48h of continuous exposure with these compounds. During the first 24h, HK-2 cells were slightly more sensitive to AL-I-NOH than AA-I, with IC50 values of 41.19 and 52.53 µM, respectively (Figure 2A; Table I). With increasing duration of exposure, AA-I caused more cytotoxicity (IC50 12.24 µM) than AL-I-NOH (IC50 21.34 µM). Indeed, between 24 and 48h, the rates of decline of ATP were greater for AA-I than for AL-I-NOH treated cells (Figure 2B and C; Table I).
Figure 2.
Cytotoxicity and genotoxicity of AA-I and AL-I-NOH in HK-2 cells. (A–F) HK-2 cells, a human proximal tubule cell line, were exposed to AA-I and AL-I-NOH under conditions described in Materials and methods. Cytotoxicity (A–C) was determined by the decline in ATP levels over time and across a dose range of compounds, compared with a DMSO-treated control. To estimate genotoxicity (D, E) of AA-I and AL-I-NOH, DNA was isolated from exposed cells and analyzed for adducts by 32P-postlabeling. (A) Varying concentrations of AA-I and AL-I-NOH in HK-2 cells exposed to the compounds. Filled (●) and open circles (○)—24h exposure to AA-I and AL-I-NOH, respectively; filled (■) and open squares (□)—48h exposure to AA-I and AL-I-NOH, respectively. Cytotoxicity (B, C) and genotoxicity (D) measured over time in HK-2 cells exposed to 5 (-●-), 12.5 (-■-) and 25 µM (-□-) of AA-I and AL-I-NOH. The compound and adducted nucleoside are indicated in corresponding panels. (E) Fragment of a 30% polyacrylamide gel after 32P-labeling of DNA adduct nucleosides. Cells were treated with 12.5 and 25 µM AA-I for 4, 8 and 24h, and the DNA was analyzed for adducts. All exposures were conducted in triplicate, digested separately and loaded into adjacent wells. St—mixture of 24-mer oligonucleotides (30fmol each) containing a single dG-AL-II or dA-AL-II, represented by the upper and lower bands, respectively. Each band corresponds to 2 adducts/106 nucleotides for 5 µg DNA. For each analysis, three standard mixtures were digested separately and loaded in adjacent wells. (F) Differential rates of accumulation of dG-AL-I and dA-AL-I in cells exposed to 5 µM of AA-I and AL-I-NOH. Rates were evaluated by using linear regression analyses in Sigma Plot. All results are shown as mean values ± SD and represent at least three independent experiments.
Table I.
IC50 values, cytotoxicity rates and AL-I-DNA adduct accumulation in HK-2 cells
| Compound | Exposure (h) | IC50 (µM) | Compound (µM) | % decline in ATP/h | AL-I-DNA/(107 nucleotides/h) |
|---|---|---|---|---|---|
| AA-I | 24 | 52.53±6.36 | 5 | NA | 1.22±0.15 |
| 12.5 | 2.04±0.10 | 5.99±1.25 | |||
| 48 | 12.24±1.39 | 25 | 2.32±0.15 | 8.93±1.56 | |
| AL-I-NOH | 24 | 41.19±3.94 | 5 | NA | 2.16±0.17 |
| 48 | 21.34±2.46 | 12.5 | 0.71±0.22 | 2.89±0.15 | |
| 25 | 1.37±0.20 | 3.90±0.81 |
NA, not applicable.
To estimate the genotoxicities of AA-I and AL-I-NOH, HK-2 cells were exposed for up to 48h to 5, 12.5 and 25 µM of the test compounds, which corresponds to a non-toxic dose, and IC50 values for AA-I and AL-I-NOH at 48h of exposure. Following exposure, DNA was isolated and analyzed for adducts using 32P-postlabeling coupled with polyacrylamide gel electrophoresis. Within the first 24h, AL-I-NOH (5 µM) generated three times more AL-I-DNA than did the same amount of AA-I, although at 12.5 and 25 µM the rates of AL-I-DNA formation were greater for AA-I than AL-I-NOH (Figure 2D and E; Table I). These results are consistent with those obtained for the cytotoxicity of the compounds, with low doses of AL-I-NOH being slightly more cytotoxic than AA-I during the first 24h of exposure (Figure 2A). Interestingly, dA-AL-I was the major adduct formed in HK-2 cells treated with AL-I-NOH, corresponding to 90% of all AL-I-DNA. In contrast, in cells exposed to AA-I, ~25% of the overall AL-I-DNA was represented by dG-AL-I (Figure 2D and E). This difference was reflected in the rates of adduct formation, shown in Figure 2F for the non-toxic dose of the compounds. Thus, for AL-I-NOH, dA-AL-I accumulated at least ten times faster than dG-AL-I. For AA-I, a 4-fold difference between two adduct types was observed. A similar tendency was noted for higher doses of the compounds (data not shown).
These results indicate that AL-I-NOH generates a response similar to AA-I in HK-2 cells. Overall, cytotoxicity and genotoxicity data were consistent and were used to estimate the effects of AA-I and AL-I-NOH exposure in the subsequent experiments.
PCP effect on AA-I and AL-I-NOH genotoxicity in HK-2 cells
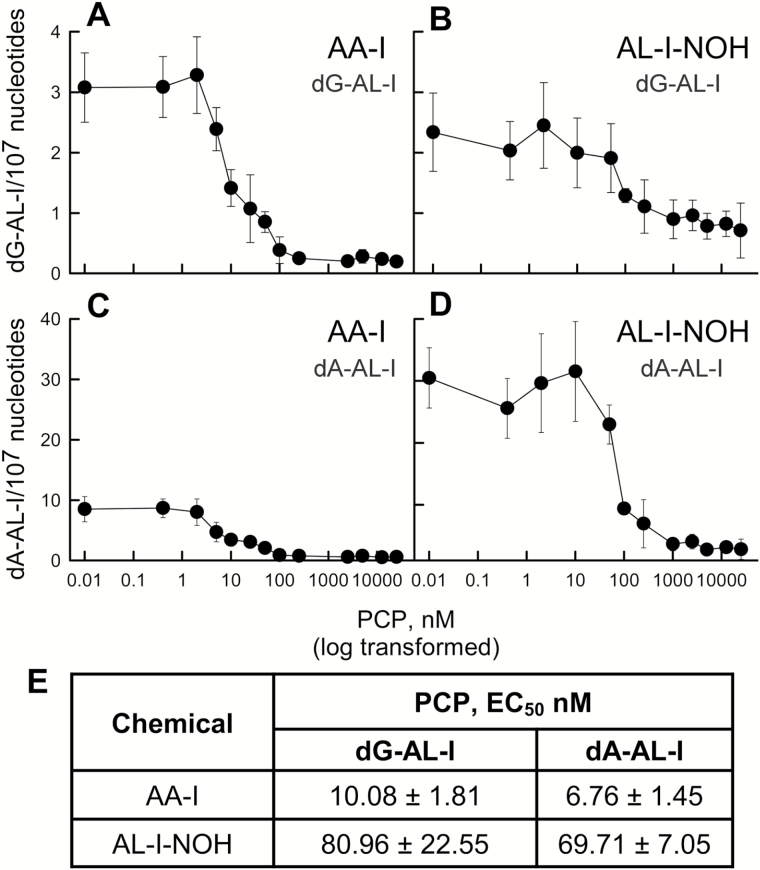
To assess the involvement of SULTs in AA-I and AL-I-NOH activation in HK-2 cells, we used PCP as an inhibitor of SULT isoforms (41,45). dG-AL-I and dA-AL-I adduct formation was monitored in DNA obtained from cells treated for 6h with 25 µM of the test compound, in the absence or the presence of 0.4–25000nM PCP (Figure 3A–D). PCP (2.5–25 µM) was not toxic to cells for up to 24h; after 48h, there was a 35% decrease in ATP levels for all PCP concentrations used, compared with a mock control (data not shown). PCP, in the nanomolar range, caused a significant decrease in dG-AL-I and dA-AL-I formation induced by AA-I and AL-I-NOH. The dependence of AL-I-DNA levels on the concentration of PCP allowed EC50 for PCP to be evaluated (Figure 3E). For AA-I, when compared with AL-I-NOH, lower EC50 values were found, namely, 10.08nM for dG-AL-I and 6.76nM for dA-AL-I. Higher PCP concentrations caused a half-maximum effect on AL-I-DNA in cells exposed to AL-I-NOH, 80.96 and 69.71nM for dG-AL-I and dA-AL-I, respectively.
Figure 3.
Effect of PCP on the genotoxicity of AA-I and AL-I-NOH in HK-2 cells. HK-2 cell cultures were exposed to 12.5 µM of AA-I or AL-I-NOH, with or without PCP, 0.4–25000nM. After 6h, cells were collected and DNA prepared for adduct analysis as described in Materials and methods. Panels (A) and (B) show the dependence of dG-AL-I formation on varying PCP concentrations in cells treated with AA-I and AL-I-NOH, respectively. Panels (C) and (D) display accumulation of dA-AL-I in cells exposed to AA-I and AL-I-NOH, respectively. Each point represents the mean value and SD for at least three independent experiments. (E) Half-inhibitory values (EC50) were calculated for the PCP effect on dG-AL-I and dA-AL-I adduct formation in HK-2 cells exposed to AA-I and AL-I-NOH. Values were obtained, using logistic regression analysis in Sigma Plot and shown as mean values and SD.
PCP effect on AA-I and AL-I-NOH cytotoxicity in HK-2 cells
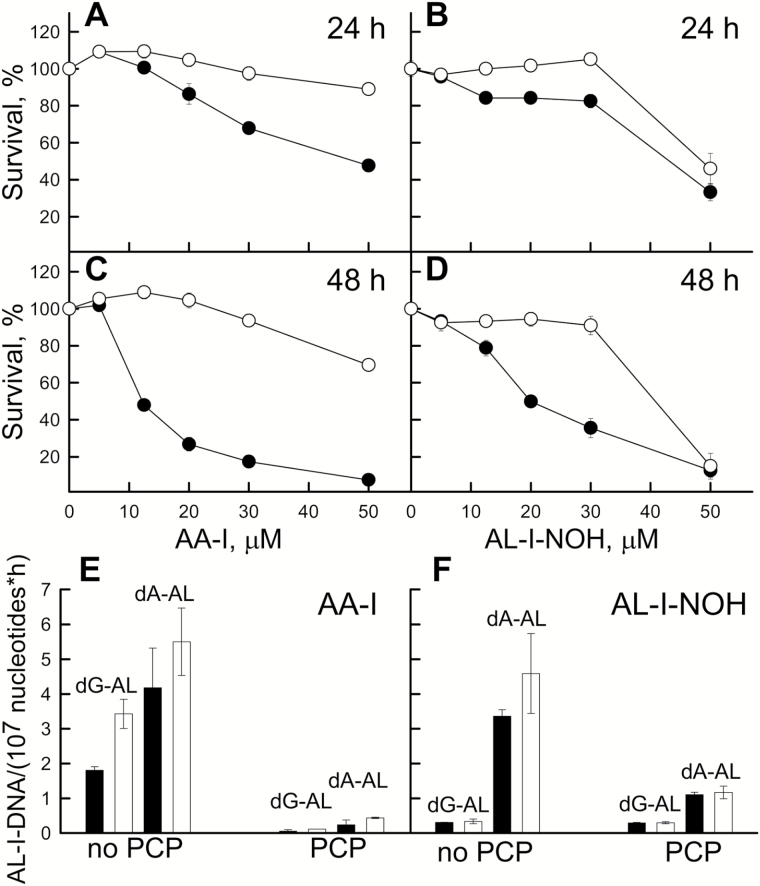
As AL-I-DNA adduct formation reflects genotoxicities of AA-I and AL-I-NOH, we examined the effect of PCP on the cytotoxicity of these compounds. Cell survival was monitored 24 and 48h after exposure to 10–50 µM of AA-I or AL-I-NOH, with or without PCP, 250nM for AA-I and 1000nM for AL-I-NOH (Figure 4A–D). These PCP concentrations correspond approximately to the 10 EC50 values determined as above. PCP alone reduced ATP levels in HK-2 cells by 10 and 20% at 24 and 48h; these levels were used as reference controls. As predicted, PCP alleviated cytoxicities caused by AA-I (Figure 4A and C) and AL-I-NOH (Figure 4B and D). This effect was more pronounced for AA-I than for AL-I-NOH. PCP failed to rescue cells when 50 µM of AL-I-NOH was present. Thus, introducing PCP at the same time as the test compounds leads to ATP levels similar to those observed for exposure to PCP alone.
Figure 4.
Effect of PCP on cytotoxicity of AA-I and AL-I-NOH in HK-2 cells. (A–D) HK-2 cells were exposed to 0–50 µM of AA-I (A and C) or AL-I-NOH (B and D) for 24 and 48h. In parallel, PCP, 250 and 1000nM, was coadministered with AA-I and AL-I-NOH, respectively. Toxicity levels were evaluated by the ATP assay. Control wells were exposed to DMSO or PCP and used as reference for cells exposed to the chemical alone (●, filled circles), or the chemical and PCP (○, empty circles), respectively. (E and F) PCP reduces rates of dG-AL-I and dA-AL-I formation caused by exposure to the compounds in HK-2 cells. Cells were treated with 12.5 (black bars) and 25 µM (white bars) of AA-I or AL-I-NOH, and AL-I-DNA levels were estimated at 6, 24, 32 and 48h after exposure. In parallel, PCP, 250 and 1000nM was coadministered with AA-I and AL-I-NOH, respectively. Rates of dG-AL-I and dA-AL-I appearance were obtained using linear regression plots in Sigma Plot. All results are shown as mean values and SD for at least three independent experiments.
Additionally, we compared the rates of AL-I-DNA formation induced by AA-I (Figure 4E) and AL-I-NOH (Figure 4F) in the presence of 250 and 1000nM PCP, respectively. PCP was more efficient in reducing rates of AL-I-DNA caused by AA-I compared with AL-I-NOH. These results correlate well with cytotoxicity studies in cells exposed to PCP and the compounds.
Gene silencing of sulfonation pathway in HK-2 and GM637 cells
To establish which SULT isoforms are involved in AA-I activation, we carried out siRNA gene knockdown experiments, selectively silencing expression of one of the following genes in HK-2 cells: SULT1A1, SULT1B1, SULT1C1, SULT1C4 and PAPSS2 (Supplementary Table S1, available at Carcinogenesis online).
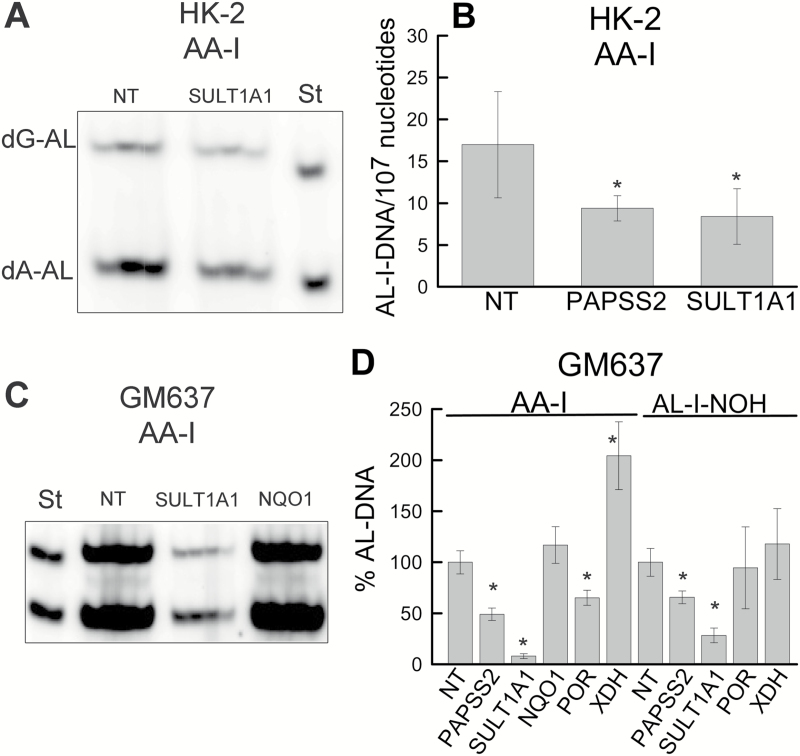
Cells exposed to NT siRNA served as a control. After 2 days of exposure to siRNAs, 25 µM AA-I was introduced for 6h, followed immediately by DNA and RNA isolation for adduct analysis and qPCR analysis, respectively. NT siRNA did not affect AL-I-DNA levels created by AA-I exposure (data not shown). Figure 5A shows a representative experiment for silencing SULT1A1 expression in HK-2 cells, demonstrating half as many AL-I-DNA adducts as in the NT control. Similar decline in AL-I-DNA formation was observed upon PAPSS2 gene silencing (Figure 5B). Both dG-AL-I and dA-AL-I were equally affected, and the combined results are reported as AL-I-DNA levels. qPCR analysis indicated that the knockdown efficiency varied significantly between cell passages, with ~1.5- and 2-fold maximum decreases observed for SULT1A1 and PAPSS2 mRNA levels, respectively (Supplementary Table S2, available at Carcinogenesis online). We did not observe a change in AL-I-DNA adduct levels in cases when SULT1B1, SULT1C2, SULT1C4 or NQO1 were targeted in HK-2 cells. These results are discussed in Supplementary Data and shown in Supplementary Figure S1A and B, available at Carcinogenesis online.
Figure 5.
Genotoxicities of AA-I and AL-I-NOH in cultured cells silenced for selected biotransformation genes of AA-I. At 30% cell density, HK-2 and GM637 cultures were treated with siRNAs for 2 and 3 days, respectively, before introducing 25 µM of one of the test compounds. Following exposure to the compound, 6h for HK-2 cells and 24h for GM637, DNA (5 µg) was subjected to adduct analysis. Panels (A) and (C) are representative fragments of two separate polyacrylamide gels showing the results of postlabeling analysis in HK-2 and GM637 cells, respectively, treated with NT, SULT1A1 or NQO1 (GM637 cells only) siRNA followed by AA-I exposure. St—mixture of standard oligonucleotides with dG-AL-II (upper band) and dA-AL-II adducts (lower band), 15fmol of each in (A) and 30fmol of each in (C). Each well represents a digestion mixture corresponding to one cell culture plate. The well between each siRNA exposure conditions and standards is left empty. Three and four digestion reactions are shown for HK-2 (A) and GM637 (C) gels, respectively. (B) AL-I-DNA adduct levels in HK-2 cells exposed to AA-I and various siRNAs. (D) AL-I-DNA levels in GM637 cells exposed to AA-I or AL-I-NOH, expressed as percentage from adduct levels data in cells treated with NT siRNA. Results are shown as the mean ± SD for at least two independent cell passages. As compared with the NT group, * indicates P < 0.05. In experiments with AA-I and AL-I-NOH, the levels of AL-I-DNA in GM637 cells treated with NT siRNA were 180±25 adducts/107 nucleotides and 235±41 adducts/107 nucleotides, respectively.
Due to the resistance of HK-2 cells to knockdown with siRNAs, other human cell lines, HEK293T17, XPA and GM637, were evaluated in a similar setting. The first two of these cell lines were sensitive to siRNA exposure and were dislodged from plates upon addition of AA-I, which prevented their use in the knockdown assay (data not shown). In contrast, the GM637 fibroblast cell line, used in our previous studies of DNA repair pathways for aristolactam-DNA adducts (21), and shown to be sensitive to AL-I-NOH exposure (43), proved eminently suitable for this analysis. Validation of knockdown efficiency by qPCR showed a decrease in mRNA levels for SULT1A1 and PAPSS2 by factors of 12.6 and 26, respectively (Supplementary Table S2, available at Carcinogenesis online).
To assess the effect of silencing of SULT1A1 and PAPSS2 on AL-I-DNA adduct formation, the GM637 cell line was exposed to NT or selected siRNA for 3 days before introducing 25 µM AA-I or AL-I-NOH for 24h. Our previous studies showed that, at this duration of exposure and dose, adduct formation in these cells is linear and cytotoxicity did not develop (21). Figure 5C shows representative 32P-postlabeling polyacrylamide gel electrophoresis adduct analysis with DNA obtained from cells, with four samples selected from two different cell passages for each condition. These cells were pre-exposed to NT or SULT1A1 siRNA, followed by AA-I treatment. SULT1A1 silencing led to a decrease in AL-I-DNA levels, by at least one order of magnitude. For AL-I-NOH, knockdown of SULT1A1 led to about a 4-fold decrease in AL-I-DNA levels (Figure 5D). Reducing the expression of PAPSS2 led to half as many adducts as in the NT control (Figure 5D) for both AA-I and AL-I-NOH.
Silencing of SULT1B1 and SULT1A2 was also tested in GM637, showing no effect or a SULT1A1 mediated decrease in AL-I-DNA levels, respectively (Supplementary Figure S1C and D, available at Carcinogenesis online).
Gene silencing of potential nitroarene reductases in GM637 cells
Since direct evidence for the involvement of nitroreductases in AA-I bioactivation in cells is lacking or controversial, we employed siRNA gene silencing approach in GM637 cells to the following genes: NQO1, POR and XDH. Both AA-I and AL-I-NOH were studied (Figure 5D). Surprisingly, despite >85% decrease in mRNA levels for NQO1, in response to transfection (Supplementary Table S2, available at Carcinogenesis online), we did not observe a significant change in AA-I genotoxicity (Figure 5C and D). Instead, the knockdown of POR led to a 50% decrease in AL-I-DNA levels compared with the NT control (Figure 5D). In addition, for AA-I, reducing the expression of XDH led to twice as many AL-I-DNA adduct as in control cells. Meanwhile, mean values of AL-I-DNA adducts generated by exposure to AL-I-NOH were not significantly affected in cells silenced for POR and XDH (Figure 5D).
Thus, POR, but not NQO1, appears to be important for the accumulation of active metabolites of AA-I, whereas XDH appears to be more proficient in detoxification rather than in activation of this toxin in cells.
Discussion
In this study, we explored the contribution of SULTs and nitroreductases to the activation of AA-I and AL-I-NOH in HK-2 and GM637 cells. Novel findings include evidence that (i) AL-I-NOH is both cytotoxic and genotoxic to HK-2 cells; (ii) AA-I and AL-I-NOH display reduced toxicities in HK-2 cells in the presence of the SULT inhibitor, PCP; (iii) silencing SULT1A1 followed by exposure to AA-I reduces AL-I-DNA adduct levels in HK-2 and GM637 cells; (iv) XDH promotes detoxification of AA-I, whereas NQO1, apparently, is not involved in bioactivation or detoxification and (v) POR promotes accumulation of active intermediates of AA-I.
The mutagenicity and toxicity of nitroaromatic compounds generally are associated with enzymatic NR (34,46). Six-electron reduction in sequential, two-electron transfer steps, affords nitroso, hydroxylamino and amino metabolites. For AA-I, the latter two intermediates correspond to AL-I-NOH and AL-I (Figure 1). Alternatively, two sequential one-electron transfers may occur, producing a nitro anion radical and a nitroso compound.
The reactivity of AA-I with DNA in the presence of nitroreductases and their cofactors, or with rat liver microsomes, is reduced in the presence of oxygen (47). However, it is unclear whether there is differential efficiency in the stepwise NR of AA-I and exactly how NR of this carcinogen occurs in cells and tissues.
NQO1 is capable of transferring two electrons and has been postulated to be the primary cytosolic nitroreductase involved in AA-I activation (36). Support for this mechanism is based largely on measurements of DNA adduct formation in in vitro reactions catalyzed by NQO1 or cytosols in the presence of various inhibitors and cofactors (48). In mice, administration of dicoumarol attenuated AA-I nephrotoxicity and decreased renal AL-I levels (37). Nevertheless, despite efficient (87%) knockdown of NQO1, AL-I-DNA adduct levels in GM637 cells exposed to AA-I remain unchanged. Similar observations were made in HK-2 cells, although knockdown efficiency could not be evaluated in this cell line (data not shown). NQO1 is present in GM637 cells (49); thus, other nitroreductases could potentially be involved in the activation of AA-I.
Knockdown of POR, the gene product of which is present in GM637 cells (49), led to a 2-fold decrease in AL-I-DNA, suggesting its potential involvement in AA-I bioactivation. POR could play two roles in AA metabolism, bioactivation or detoxification. First, POR is known for its NR capacity with various drugs, as well as being implicated in AA-I activation in vitro (36). Second, POR supports the function of various CYPs, removing AA-I available for bioactivation by producing AA-Ia (Figure 1). Since cells in culture commonly lose CYP activities and, in primary fibroblasts, minor amounts of CYP1A1, but not CYP1A2 mRNA are present (50), it appears that POR functions as a nitroreductase for AA-I in GM637 cells. Conversely, in mice, lacking POR function in liver, AA-I generated more AL-I-DNA adducts in various organs due to inhibition of CYP-driven detoxification pathway (32), implying that POR function involves more than NR of AA-I.
We observed also that XDH silencing increased AL-I-DNA levels in GM637 cells exposed to AA-I. Surprisingly, the expected decrease in adduct formation upon transformation did not occur. XDH has been proposed as a nitroreductase for AA-I (33,36). Under anaerobic conditions, incubation of XDH with AA-I and DNA lead to the formation of the non-toxic AL-I, AL-I-DNA and small quantities of 7-hydroxyaristolactam (33). The authors suggest that 7-hydroxyaristolactam is a product of intramolecular rearrangement of AL-I-NOH, concluding that AL-I-NOH is the proximate reactive metabolite of AA-I. Our results show that XDH is involved in the NR of AA-I in human cells, most likely by producing more efficiently the non-toxic AL-I (Figure 1). Conversely, as shown in this study, POR may be more efficient in supplying AL-I-NOH.
Hydroxylamine metabolites of nitroheterocyclic drugs are generally considered to be the reactive species responsible for the bactericidal effects of these drugs on anaerobic bacteria (46). Likewise, AL-I-NOH is considered to be the major precursor of reactive cyclic nitrenium species and of AL-I-DNA adducts (33,35). Recently, we reported that AL-I-NOH reacts only weakly with DNA (43). At high concentrations of this compound, and with prolonged incubation, similar levels of AL-I-DNA were observed in reactions involving AA-I, DNA and NQO1 (48). To react efficiently with DNA, AL-I-NOH requires further activation by SULT enzymes: SULT1B1 >> SULT1A1 > SULT1A2 >> SULT1A3 (43). We propose here the involvement of SULTs in AA-I activation in cells, based in part, on inhibition by PCP of AA-I and AL-I-NOH cytotoxicity and genotoxicity in human kidney cells. These results are supported by experiments in which the SULT1A1 gene was silenced, revealing a major decrease in AL-I-DNA adduct levels in GM637 and, to a lesser extent, HK-2 cells exposed to AA-I. For many carcinogens, the ultimate carcinogenic species are represented by N-hydroxy metabolites and their sulfuric acid esters (40). Pretreatment of rodents exposed to N-hydroxy-2-acetylaminofluorene with PCP inhibits its sulfonation, thus, preventing DNA adduct and tumor formation in hepatic tissues. The role of human SULTs in bioactivation of xenobiotics, and, in particular, nitroaromatic compounds including AA-I, has been studied by Glatt et al. (39) using mutagenesis assays in Salmonella and mammalian cells expressing various human SULTs. Our studies are consistent with their report showing that the expression of human SULT1A1 in Salmonella increases the mutagenicity of AA-I (41).
We were unable to amplify the transcript of SULT1B1 in HK-2 or GM637 cells; accordingly, we cannot confirm the effects of its knockdown on AL-I-DNA adduct formation in these cell lines. Since SULT1B1 is proficient in AL-I-NOH sulfonation (42), it is unclear whether this SULT is involved in AA-I bioactivation in vivo, especially if activation occurs primarily in the liver, with active metabolites being transported in the blood to target tissues, as proposed for methylpyrene and its hydroxy and sulfonyloxy metabolites (51). Finally, the potential roles of SULT1E1 and SULT2A1, abundant in human tissues (52), cannot be disregarded.
Intriguing, we found that the fold-difference between dA-AL-I and dG-AL-I adduct levels is much higher in AL-I-NOH than in AA-I treated cells. Clues for the mechanism involved can be obtained from in vitro experiments in which more dA-AL-I than dG-AL-I adduct was detected in DNA exposed to AL-I-N-OSO3, either synthetic or produced by the activation of AL-I-NOH by SULTs and PAPS (43). Interestingly, when AA-I is activated by nitroreductases only or non-enzymatic zinc reduction, comparable levels of dA-AL-I and dG-AL-I adducts are detected. A possible explanation for this result would be that AL-I-N-OSO3 prefers non-covalent binding in vicinity to dA residues in DNA. Thus, when cells exposed to AL-I-NOH, the compound receives additional activation through sulfonation and, as a consequence, preferential formation of dA-AL-I occurs. In the case where AA-I is introduced into cells, certain nitroreductases, upon AA-I transformation into AL-I-NOH, may remain bound to its product and delay or prevent its sulfonation to some extent, thus leading to more dG-AL-I compared with cells treated with AL-I-NOH.
Thus, our results suggest that, in vivo, dA-AL-I adduct is mainly formed through the Phase II activation of AL-I-NOH, whereas dG-AL-I adduct may appear through the same mechanism or Phase I activation of AA-I. Observed higher levels of dA-AL-I than dG-AL-I in individuals exposed to AA-I provide additional support to the importance of sulfonation for the activation of this carcinogen in human body. Previously, this difference was mainly attributed to the deficiency for the global genomic repair of dA-AL-I (21). According to the results reported in this study, the efficient repair of dG-AL-I and preferential activation of AA-I through the sulfonation in vivo may account for this observation.
In summary, our studies show that SULTs, in particular SULT1A1, are essential for the DNA adduct formation mediated by AA-I, very likely, by participating in the sulfonation of its partial NR product, AL-I-NOH. A search for nitroreductases acting on AA-I revealed that XDH is more efficient in detoxification than in bioactivation of AA-I, whereas POR is important for AA-I activation. Importantly, our results suggest that NQO1 does not play a critical role in AA-I metabolism in the cell lines studied here.
Supplementary material
Supplementary Material can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (ES004068 to A.P.G.); Henry and Marsha Laufer.
Supplementary Material
Acknowledgements
The authors thank K.G.Dickman and her colleagues for sharing unpublished microarray data for HK-2 cells.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AA
aristolochic acid
- AL-II-NOH
N-hydroxyaristolactam II
- AL-I-NOH
N-hydroxyaristolactam I
- AL-I-N-OSO3H
aristolactam-I-N-sulfate
- dA-AL-I
7-(deoxyadenosin-N6-yl)-aristolactam I
- dA-AL-II
7-(deoxyadenosin-N6-yl)-aristolactam II
- dG-AL-I
7-(deoxyguanosin-N2-yl)-aristolactam I
- dG-AL-II
7-(deoxyguanosin-N2-yl)-aristolactam II
- DMSO
dimethyl sulfoxide
- NQO1
NAD(P)H:quinone oxidoreductase 1
- NR
nitroreduction
- NT
non-target siRNA
- PAPSS2
3′-phosphoadenosine-5′-phosphosulfate synthase 2
- PCP
pentachlorophenol
- POR
cytochrome P450 oxidoreductase
- qPCR
quantitative PCR
- SULT
sulfotransferase
- UTUC
upper urinary tract urothelial carcinoma
- XDH
xanthine dehydrogenase
References
- 1. Vanherweghem J.L., et al. (1993) Rapidly progressive interstitial renal fibrosis in young women: association with slimming regimen including Chinese herbs. Lancet, 341, 387–391. [DOI] [PubMed] [Google Scholar]
- 2. Nortier J.L., et al. (2000) Urothelial carcinoma associated with the use of a Chinese herb (Aristolochia fangchi). N. Engl. J. Med., 342, 1686–1692. [DOI] [PubMed] [Google Scholar]
- 3. Cosyns J.P., et al. (1994) Chinese herbs nephropathy: a clue to Balkan endemic nephropathy? Kidney Int., 45, 1680–1688. [DOI] [PubMed] [Google Scholar]
- 4. Grollman A.P., et al. (2007) Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl Acad. Sci. USA, 104, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jelaković B., et al. (2012) Aristolactam-DNA adducts are a biomarker of environmental exposure to aristolochic acid. Kidney Int., 81, 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vanhaelen M., et al. (1994) Identification of aristolochic acid in Chinese herbs. Lancet, 343, 174. [DOI] [PubMed] [Google Scholar]
- 7. Gillerot G., et al. (2001) Aristolochic acid nephropathy in a Chinese patient: time to abandon the term “Chinese herbs nephropathy”? Am. J. Kidney Dis., 38, E26. [DOI] [PubMed] [Google Scholar]
- 8. De Broe M.E. (2012) Chinese herbs nephropathy and Balkan endemic nephropathy: toward a single entity, aristolochic acid nephropathy. Kidney Int., 81, 513–515. [DOI] [PubMed] [Google Scholar]
- 9. Kumar V., et al. (2003) Naturally occurring aristolactams, aristolochic acids and dioxoaporphines and their biological activities. Nat. Prod. Rep., 20, 565–583. [DOI] [PubMed] [Google Scholar]
- 10. Vaclavik L., et al. (2014) Quantification of aristolochic acids I and II in herbal dietary supplements by ultra-high-performance liquid chromatography-multistage fragmentation mass spectrometry. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess., 31, 784–791. [DOI] [PubMed] [Google Scholar]
- 11. Mengs U., et al. (1982) The carcinogenic action of aristolochic acid in rats. Arch. Toxicol., 51, 107–119. [Google Scholar]
- 12. Mengs U. (1988) Tumor-induction in mice following exposure to aristolochic acid. Arch. Toxicol., 61, 504–505. [DOI] [PubMed] [Google Scholar]
- 13. Cosyns J.P., et al. (2001) Chronic aristolochic acid toxicity in rabbits: a model of Chinese herbs nephropathy? Kidney Int., 59, 2164–2173. [DOI] [PubMed] [Google Scholar]
- 14. Schmeiser H.H., et al. (1984) Mutagenicity of the two main components of commercially available carcinogenic aristolochic acid in Salmonella typhimurium . Cancer Lett., 23, 97–101. [DOI] [PubMed] [Google Scholar]
- 15. Schmeiser H.H., et al. (1990) Aristolochic acid activates ras genes in rat tumors at deoxyadenosine residues. Cancer Res., 50, 5464–5469. [PubMed] [Google Scholar]
- 16. Sato N., et al. (2004) Acute nephrotoxicity of aristolochic acids in mice. J. Pharm. Pharmacol., 56, 221–229. [DOI] [PubMed] [Google Scholar]
- 17. Shibutani S., et al. (2007) Selective toxicity of aristolochic acids I and II. Drug Metab. Dispos., 35, 1217–1222. [DOI] [PubMed] [Google Scholar]
- 18. Pfau W., et al. (1990) 32P-postlabelling analysis of the DNA adducts formed by aristolochic acid I and II. Carcinogenesis, 11, 1627–1633. [DOI] [PubMed] [Google Scholar]
- 19. Dong H., et al. (2006) Quantitative determination of aristolochic acid-derived DNA adducts in rats using P-32-postlabeling/polyacrylamide gel electrophoresis analysis. Drug Metab. Dispos., 34, 1122–1127. [DOI] [PubMed] [Google Scholar]
- 20. Attaluri S., et al. (2010) DNA adducts of aristolochic acid II: total synthesis and site-specific mutagenesis studies in mammalian cells. Nucleic Acids Res., 38, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sidorenko V.S., et al. (2012) Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res., 40, 2494–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C.H., et al. (2012) Aristolochic acid-associated urothelial cancer in Taiwan. Proc. Natl Acad. Sci. USA, 109, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoang M.L., et al. (2013) Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med., 5, 197ra102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poon S.L., et al. (2013) Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci. Transl. Med., 5, 197ra101. [DOI] [PubMed] [Google Scholar]
- 25. Vogelstein B., et al. (2013) Cancer genome landscapes. Science, 339, 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scelo G., et al. (2014) Variation in genomic landscape of clear cell renal cell carcinoma across Europe. Nat. Commun., 5, 5135. [DOI] [PubMed] [Google Scholar]
- 27. Grollman A.P. (2013) Aristolochic acid nephropathy: harbinger of a global iatrogenic disease. Environ. Mol. Mutagen., 54, 1–7. [DOI] [PubMed] [Google Scholar]
- 28. Rosenquist T.A., et al. (2010) Cytochrome P450 1A2 detoxicates aristolochic acid in the mouse. Drug Metab. Dispos., 38, 761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shibutani S., et al. (2010) Detoxification of aristolochic acid I by O-demethylation: less nephrotoxicity and genotoxicity of aristolochic acid Ia in rodents. Int. J. Cancer, 127, 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chan W., et al. (2006) Study of the phase I and phase II metabolism of nephrotoxin aristolochic acid by liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom., 20, 1755–1760. [DOI] [PubMed] [Google Scholar]
- 31. Krumbiegel G., et al. (1987) Studies on the metabolism of aristolochic acids I and II. Xenobiotica, 17, 981–991. [DOI] [PubMed] [Google Scholar]
- 32. Xiao Y., et al. (2008) Hepatic cytochrome P450s metabolize aristolochic acid and reduce its kidney toxicity. Kidney Int., 73, 1231–1239. [DOI] [PubMed] [Google Scholar]
- 33. Pfau W., et al. (1990) Aristolochic acid binds covalently to the exocyclic amino group of purine nucleotides in DNA. Carcinogenesis, 11, 313–319. [DOI] [PubMed] [Google Scholar]
- 34. Boelsterli U.A., et al. (2006) Bioactivation and hepatotoxicity of nitroaromatic drugs. Curr. Drug Metab., 7, 715–727. [DOI] [PubMed] [Google Scholar]
- 35. Schmeiser H.H., et al. (2009) Chemical and molecular basis of the carcinogenicity of Aristolochia plants. Curr. Opin. Drug Discov. Dev., 12, 141–148. [PubMed] [Google Scholar]
- 36. Stiborová M., et al. (2013) Enzymes metabolizing aristolochic acid and their contribution to the development of aristolochic acid nephropathy and urothelial cancer. Curr. Drug Metab., 14, 695–705. [DOI] [PubMed] [Google Scholar]
- 37. Chen M., et al. (2011) Inhibition of renal NQO1 activity by dicoumarol suppresses nitroreduction of aristolochic acid I and attenuates its nephrotoxicity. Toxicol. Sci., 122, 288–296. [DOI] [PubMed] [Google Scholar]
- 38. Stiborová M., et al. (2014) The influence of dicoumarol on the bioactivation of the carcinogen aristolochic acid I in rats. Mutagenesis, 29, 189–200. [DOI] [PubMed] [Google Scholar]
- 39. Glatt H. (2000) Sulfotransferases in the bioactivation of xenobiotics. Chem. Biol. Interact., 129, 141–170. [DOI] [PubMed] [Google Scholar]
- 40. Miller J.A. (1994) Recent studies on the metabolic activation of chemical carcinogens. Cancer Res., 54, 1879s–1881s. [PubMed] [Google Scholar]
- 41. Meinl W., et al. (2006) Human sulphotransferases are involved in the activation of aristolochic acids and are expressed in renal target tissue. Int. J. Cancer, 118, 1090–1097. [DOI] [PubMed] [Google Scholar]
- 42. Stiborová M., et al. (2011) The human carcinogen aristolochic acid i is activated to form DNA adducts by human NAD(P)H:quinone oxidoreductase without the contribution of acetyltransferases or sulfotransferases. Environ. Mol. Mutagen., 52, 448–459. [DOI] [PubMed] [Google Scholar]
- 43. Sidorenko V.S., et al. (2014) Bioactivation of the human carcinogen aristolochic acid. Carcinogenesis, 35, 1814–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Attaluri S., et al. (2014) Total synthesis of the aristolochic acids, their major metabolites, and related compounds. Chem. Res. Toxicol., 27, 1236–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L.Q., et al. (2006) Inhibition of sulfotransferases by xenobiotics. Curr. Drug Metab., 7, 83–104. [DOI] [PubMed] [Google Scholar]
- 46. Kedderis G.L., et al. (1988) The metabolic activation of nitroheterocyclic therapeutic agents. Drug Metab. Rev., 19, 33–62. [DOI] [PubMed] [Google Scholar]
- 47. Schmeiser H.H., et al. (1997) Comparison of DNA adduct formation by aristolochic acids in various in vitro activation systems by 32P-post-labelling: evidence for reductive activation by peroxidases. Carcinogenesis, 18, 1055–1062. [DOI] [PubMed] [Google Scholar]
- 48. Stiborová M., et al. (2002) Carcinogenic aristolochic acids upon activation by DT-diaphorase form adducts found in DNA of patients with Chinese herbs nephropathy. Carcinogenesis, 23, 617–625. [DOI] [PubMed] [Google Scholar]
- 49. Prins J.M., et al. (2014) Cd2+-induced alteration of the global proteome of human skin fibroblast cells. J. Proteome Res., 13, 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saeki M., et al. (2002) mRNA expression of multiple cytochrome p450 isozymes in four types of cultured skin cells. Int. Arch. Allergy Immunol., 127, 333–336. [DOI] [PubMed] [Google Scholar]
- 51. Ma L., et al. (2003) Albumin strongly prolongs the lifetime of chemically reactive sulphuric esters and affects their biological activities in the rat. Nova Acta Leopold., 329, 265–272. [Google Scholar]
- 52. Riches Z., et al. (2009) Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab. Dispos., 37, 2255–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.