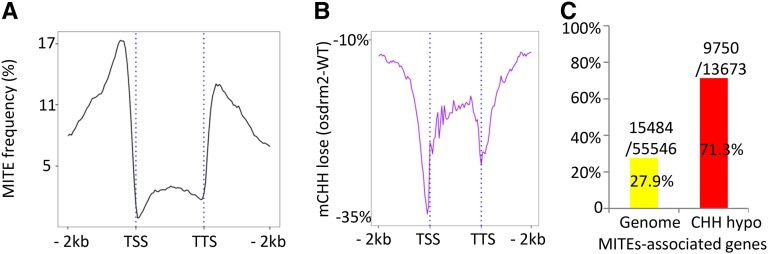
Figure 4.
The osdrm2 mutation reduces CHH methylation of MITEs that are located close to genes. A, Frequency of the occurrence of MITEs in rice protein-coding genes. The rice protein-coding genes (including the body and the 2-kb upstream and downstream regions) were divided into 120 intervals (x axis) and searched for the presence of MITE sequences at each interval. The y axis shows the percentage of rice genes with the presence of MITEs. B, Distribution of genome-wide average loses of CHH methylation in the rice protein-coding genes in ordrm2. All rice protein-coding genes (including the body and the 2-kb upstream and downstream regions) were divided into 120 intervals (x axis), and CHH methylation losses at each interval in osdrm2 compared with the wild type (WT) are shown as minus percentages (y axis). C, High enrichment of MITE-associated genes in hypo-CHH methylation (71.4%) in osdrm2, compared with 27.9% of MITE-associated genes genome wide. TSS, Transcription start site; TTS: transcription terminal site.