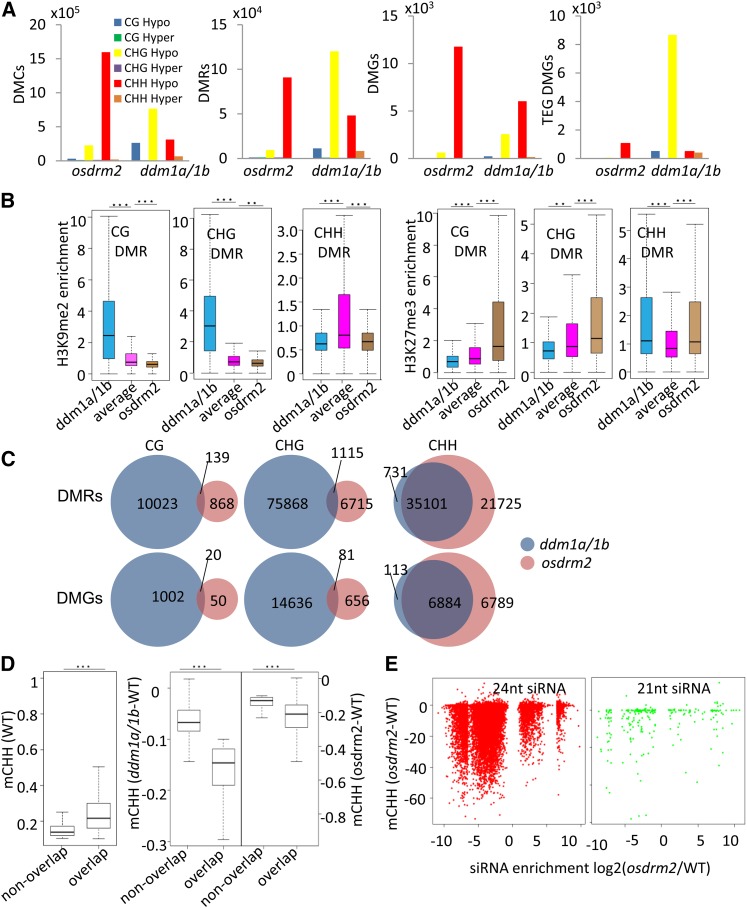
Figure 5.
Analysis of DMCs, DMRs, and DMGs detected in osddm1a/1b and osdrm2 mutants. A, Numbers of hypo- and hyper-DMCs/DMRs/DMGs detected in osddm1a/1b and osdrm2. DMGs of protein-coding genes and TEGs are shown. B, Enrichment of H3K9me2 or H3K27me3 of hypo-CG, -CHG, and -CHH DMRs in osddm1a/1b and osdrm2 plants relative to genome-wide averages. Significant enrichments are indicated by double asterisks (P < 0.01) or triple asterisks (P < 0.001, Student’s t test). C, Venn diagrams of hypo-CG, -CHG, and -CHH DMRs and DMGs in osddm1a/1b and osdrm2. D, Box plots of CHH methylation levels of overlapped and nonoverlapped hypo-CHH DMRs in wild-type (WT) plants (left) and CHH methylation losses of overlapped hypo-CHH DMRs and nonoverlapped DMRs in osdrm2 and osddm1a/1b plants (right). Significant differences are indicated by triple asterisks (P < 0.001, Student’s t test). E, CHH methylation level changes (y axis; in minus percentage) of each bin (1 kb) with differential 24- and 21-nucleotide siRNA levels (x axis; false discovery rate < 0.01, Student’s t test) in osdrm2 compared with the wild type.