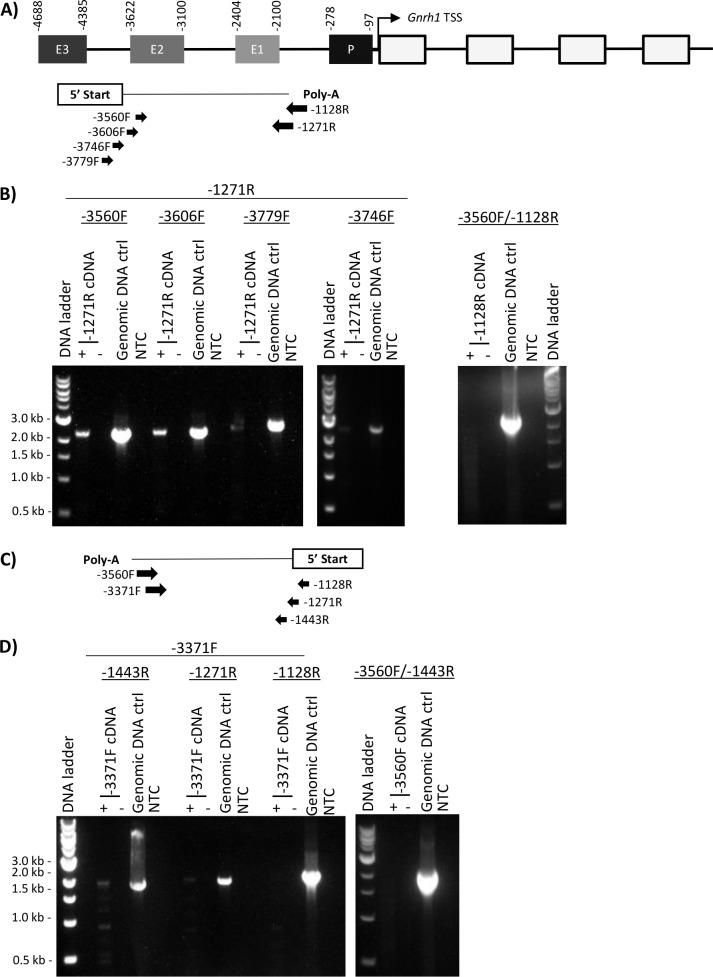
Fig 3. Strand-specific cDNA and RT-PCR analysis of the mouse GnRH-E1 RNA variants.
(A) Schematic diagram of the sense GnRH-E1 RNA variant, with a 3’ polyA site located downstream of GnRH-E1 as predicted by RACE. PCR primers in reverse direction at -1128 bp or -1271 bp (reverse arrows) were used for strand-specific cDNA synthesis to capture the sense GnRH-E1 RNA. PCR analysis was performed using -1271 bp or -1128 bp reverse primer paired with the forward primers at -3560 bp, -3606 bp, -3779 bp, and -3746 bp (forward arrows) from the mouse Gnrh1 TSS. Primer positions are aligned to the mouse conserved regulatory elements and coordinates diagrammed above. (B) Strand-specific cDNA synthesized using the -1271 bp reverse primer was subject to PCR analysis using the following primer pairs: -3560F/-1271R, -3606F/-1271R, -3779F/-1271R, -3746F/-1271R. Strand-specific cDNA synthesized using -1128 bp reverse primer was subject to PCR analysis using the primer pair -3560F/-1128R. (C) Schematic diagram of the mouse antisense GnRH-E1 RNA variant, with a 3’ polyA site predicted upstream of GnRH-E2 by RACE. PCR primers in the forward direction at -3560 bp or -3371 bp (forward arrows) was used for strand-specific cDNA synthesis to capture the antisense GnRH-E1 RNA variant. PCR analysis was performed using -3371 bp or -3560 bp forward primer paired with the reverse primers at -1443 bp, -1271 bp, -1128 bp from the Gnrh1 TSS. D) Strand-specific cDNA synthesized using the -3371 bp forward primer was subject to PCR analysis using the following PCR primer pairs -3371F/-1443R, -3371F/-1271R, and -3371F/-1128R. Strand-specific cDNA synthesized using the -3560 bp forward primer was subject to PCR analysis using the primer pair at -3560F/-1443R. All reverse transcription reactions were performed on total RNA samples with (+) and without (-) reverse transcriptase and were amplified by PCR in parallel with GT1-7 genomic DNA control and no-template water control (NTC). The size of PCR amplicons was marked by 1 kbp DNA ladder.