Figure 3.
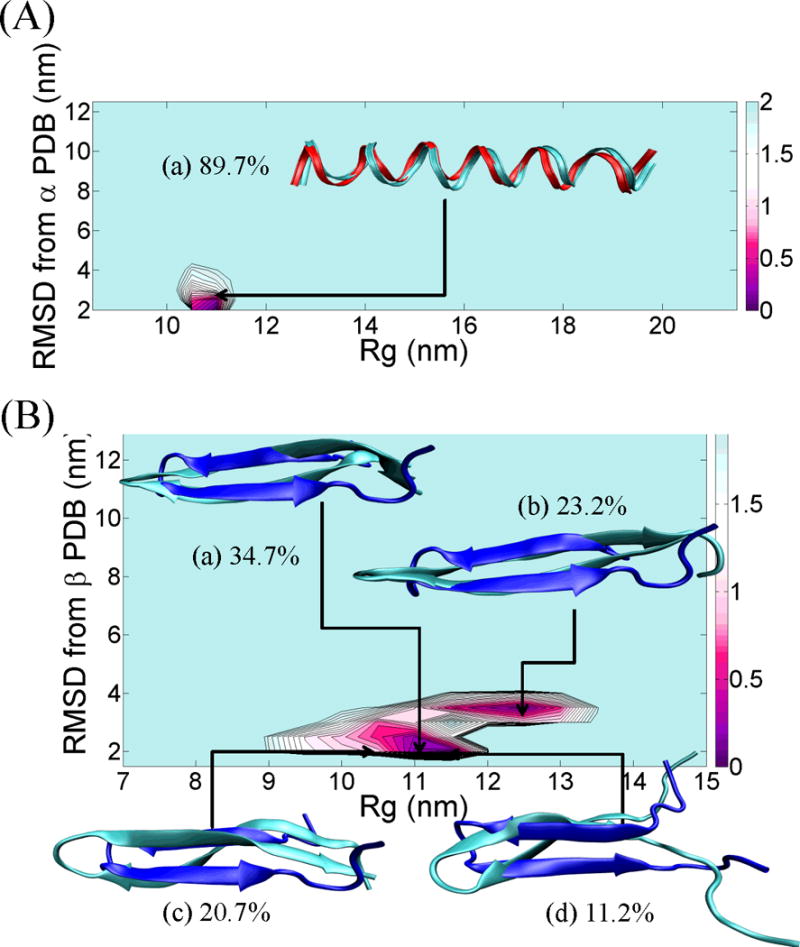
(A) PMF landscape of the chameleon- HS-α sequence fragment along with the result of cluster analysis. X-axis of the PMF graph is the radius of gyration, Rg (nm) and Y-axis of the PMF graph is the RMSD (nm) from the reference α-helix fragment of the protein, PDB id = 1JIG. The structure (a) reflected the lowest RMSD structure from each centroid and ordered by population. The red ribbon structure indicates the reference α-helix structure. (B) PMF landscape of the chameleon- HS-β sequence fragment along with the result of cluster analysis. X-axis of the PMF graph is the radius of gyration, Rg (nm) and Y-axis of the PMF graph is the RMSD (nm) from the reference β-strand fragment of the protein, PDB id = 1P35. Each of the (a), (b), (c) and (d) structures reflected the lowest RMSD structure from each centroid and ordered by population. The blue ribbon structure indicates the reference β-strand structure. All ribbon structures were prepared using VMD.42 The energy unit on this PMF landscape is kcal/mol.
