Figure 2.
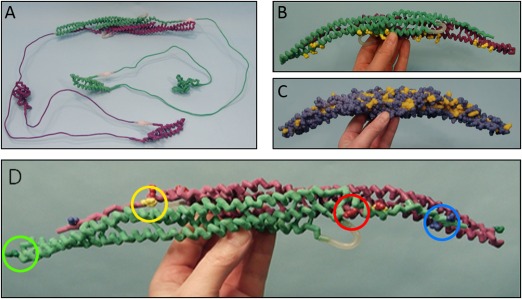
Physical protein models. The four hand‐held physical models used in 4 of the 7 discussion sections (“physical model” treatment groups). Purple and light green backbones represent separate identical monomers connected by magnets to create the CIP4 dimer. (A) Entire CIP4 dimer including FBAR (alpha helix), HR1 (alpha helix) and SH3 domains (beta sheet), made of nylon. Thin metal connections represent uncrystallized regions of the protein. Their length accurately represents the number of amino acids known to span these regions. (B ) F‐BAR dimer of CIP4, made of nylon. The positively charged amino acid side chains (lysine and arginine) important for binding to negatively charged phospholipid head groups in the membrane are displayed on the F‐BAR domain in yellow. (C ) F‐BAR dimer of CIP4 in spacefill, made of plaster. Yellow amino acid residues are hydrophobic and blue amino acid residues are hydrophilic. (D ) F‐BAR dimer of CIP4, made of nylon. The yellow amino acid visible on the purple F‐BAR monomer (the other yellow amino acid is not visible) is the hydrophobic phenylalanine (F276) residue involved in lateral interactions of CIP4 dimers. Red amino acids are the positively charged lysine residues (K273 and K66) involved in ionic interactions with the negatively charged blue amino acids glutamate (E285) and aspartate (D286) of an adjacent CIP4 dimer. The dark green amino acids at the end of each monomer are the positively charged lysine (K166) residues involved in end‐to‐end interactions of CIP4 dimers.
