Figure 5.
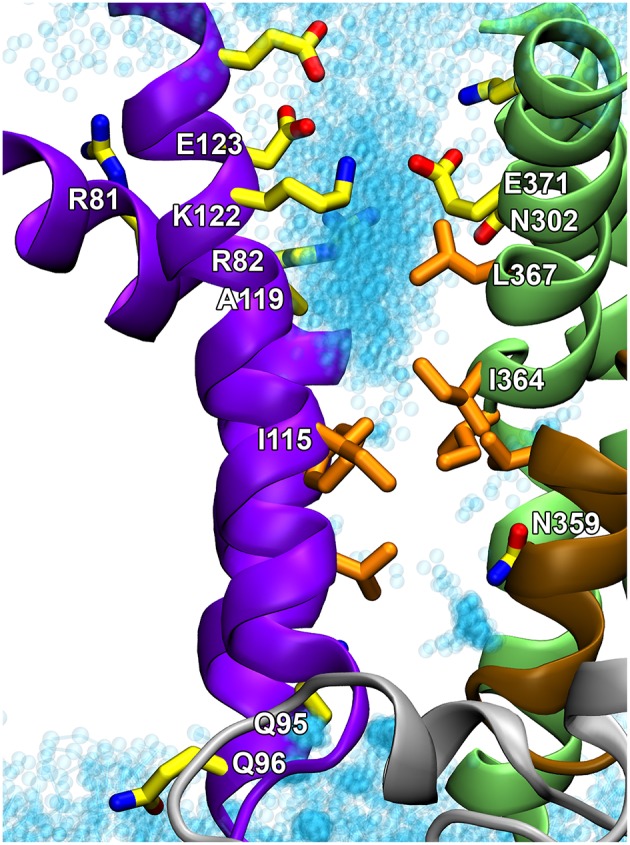
Proposed hydrophobic gate pathway in side view with indication of residues that might play a role in the recognition and translocation of the phospholipid. The E2 structural model of the ATP8A2 membrane domain (Vestergaard et al., 2014) is visualized with the cytoplasmic side up. Selected residues, some of which are mentioned in the text, are shown in licorice representation with carbons in orange for hydrophobic residues and in yellow for polar/charged residues. Water molecules in the groove are shown as cyan transparent spheres. I364 and I115 are key residues in the hydrophobic gate. N359 is important for the affinity for the lipid substrate (Vestergaard et al., 2014). Q95, Q96, A119, E123, N302, and L367 are located at positions corresponding to the yeast Dnf1 flippase residues G230, A231, T254, D258, N550, and Y618, which have been shown to be crucial to the specific lipid selectivity of this flippase (Baldridge and Graham, 2012, 2013).
