Figure 1.
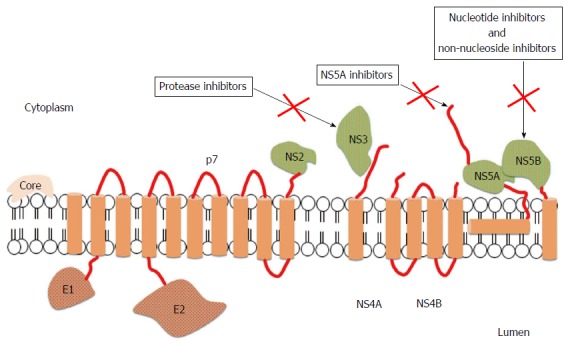
Structure of hepatitis C virus and mechanisms of action of direct antiviral agents[29]. The open reading frame of hepatitis C virus (HCV) encodes 11 proteins: 3 structural proteins (core, E1, and E2); 6 nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A and NS5B); the p7 protein; and the frameshift (F) protein (not illustrated). The core protein has a role in viral capsid formation and can directly interact with many cellular proteins and pathways implicated in the HCV lifecycle. Envelope glycoproteins, E1 and E2, are fundamental components of the virion envelope and are essential for HCV entry and fusion; in particular, E2 has a central role in the early steps of infection because it functions as a host receptor binding protein and mediates the attachment to host cells. The F protein is probably produced during viral infection and could be involved in viral persistence, but the exact role has not been fully elucidated. The p7 protein is an integral membrane protein belonging to the viroporin family and maybe acts as a calcium ion channel, but further studies are needed to confirm its function. NS2 is a non-glycosylated transmembrane protein having a protease activity, which may interact with host cell proteins. NS3 is a multi-functional protein owning a serine protease domain and a helicase/nucleoside triphosphatase domain, while NS4A is a cofactor of NS3 protease activity. The 2 proteins can interact with host cell pathways and proteins involved in HCV lifecycle and for this reason they are an appealing viral target for anti-HCV therapies. NS4 is an integral membrane protein serving as a membrane anchor for the replication complex; moreover, it can inhibit cellular syntesis and modulate the HCV RdRp activity. NS5A is a zinc-metalloprotein playing a role in virus replication, cell growth replication, and in mediating interferon-resistance, even if some of these functions need to be still clarified. NS5B belongs to the class of tail-anchored proteins. Its crystal structure showed that the RdRp has a "fingers, palm and thumb" structure; interactions between the fingers and thumb subdomains create a fully surrounded catalytic site ensuring both HCV RNA strand synthesis. For this reason, the RdRp is an attractant target for new anti-HCV drugs.
