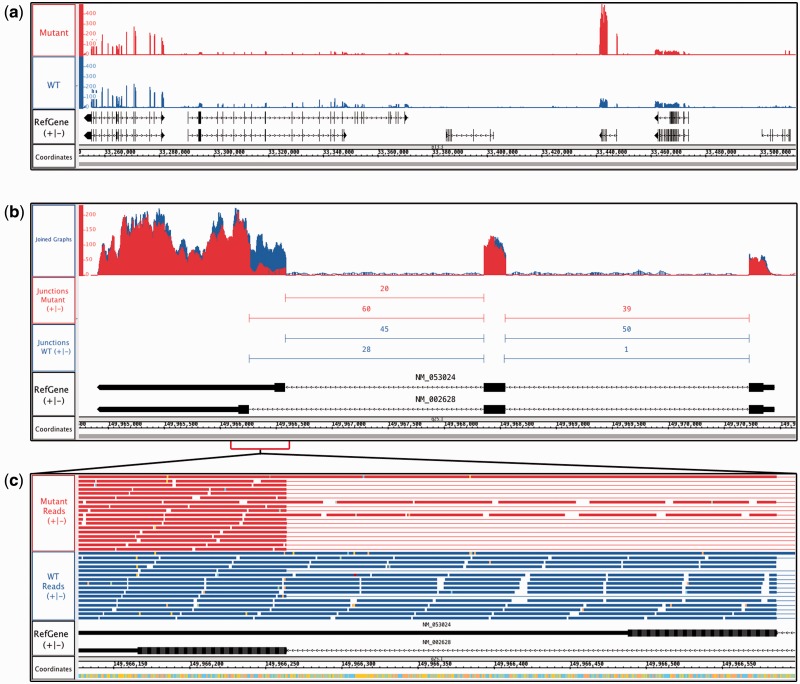
Fig. 2.
RNA-Seq data from human lung adenocarcinomas bearing mutant or wild-type (WT) alleles of the KRAS oncogene. (a) Coverage depth graphs show transcript abundance across a 250 kb region. Mutant samples contain a peak indicating higher expression in the mutant sample. (b) Overlaid depth graphs showing a discontinuity in coverage indicating differential splicing in PFN2. Quantification of split reads by FindJunctions further supports differential splicing. (c) Zoomed in view of (b), showing aligned reads