Fig. 1.
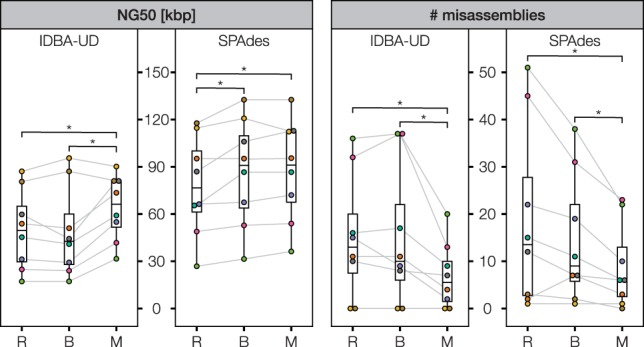
Effect on SAG assembly. We corrected the raw reads (R) with BayesHammer (B; Nikolenko et al., 2013) or MeCorS (M). We then used IDBA-UD (Peng et al., 2012) and SPAdes (Bankevich et al., 2012) to assemble the SAGs. Brackets indicate all statistically significant changes (P < 0.05; two-tailed Wilcoxon signed-rank test). Quality assessment with QUAST (Gurevich et al., 2013); Supplementary Tables S4 and S5 contain in-depth assembly statistics
