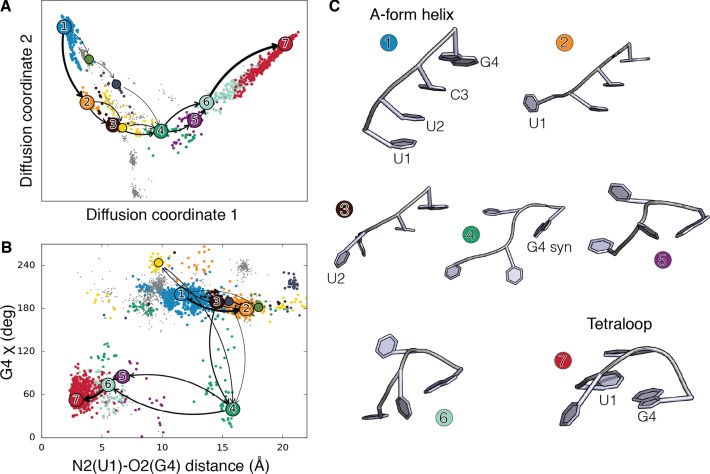
Figure 2.
(A) UUCG Helix to loop folding pathways projected on the first two diffusion coordinates. Clusters corresponding to A-form helix and to the loop are colored in blue and red, respectively. On-pathways are shown in colors, while gray points correspond to clusters not contributing to the folding pathway. Arrows show the dominant folding pathways (≈75% of the total folding flux). Line width is proportional to the flux. (B) Helix to loop folding pathway projected on the distance between atom O2 in base U1 and atom N1 in base G4 versus the glycosidic torsion angle in G4. The distance reports on the formation of a signature interaction of the tetraloop. (C) Centers of the clusters in the main folding pathway, numbered and colored as in panels A and B.