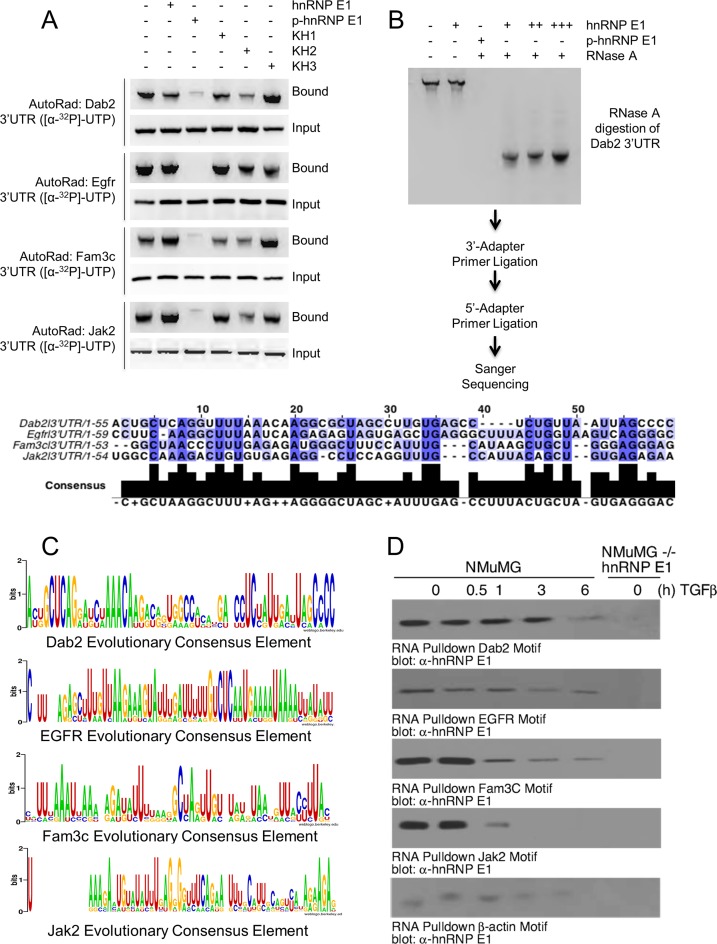
Figure 2.
EMT-inducing genes bind hnRNP E1 through their 3′-UTR and are protected from exonuclease degradation. (A) Autoradiograph analysis of RNA bound to hnRNP E1. Rows marked with ‘Bound’ contain sample that was obtained through elution of recombinant protein by reduced glutathione elution buffer. Rows marked ‘Input’ contain sample that was obtained by aliquoting a portion of the reaction prior to elution to ensure equal loading of radioactive RNA. (B) Autoradiograph of exonuclease treated samples that were bound with either hnRNP E1 or p-hnRNP E1, titration is indicated by additional ‘+’ characters. Sanger sequencing of digested fragments protected by hnRNP E1 are shown in alignment below autoradiograph. (C) Evolutionary conservation analysis of sequences obtained by Sanger. Homologene (NCBI) database was used to obtain orthologous gene sequences for evolutionarily divergent organisms. These sequences were aligned by Clustal-omega using the fragment sequence as an alignment template. Height of nucleic acid base in consensus logo indicates its degree of conservation. (D) Immunoblot analysis of hnRNP E1 pulled down by synthetic RNA for each candidate gene from TGFβ (up to 24 h) treated lysate. The data presented are typical of three independent experiments demonstrating similar loss and gain of interactions.