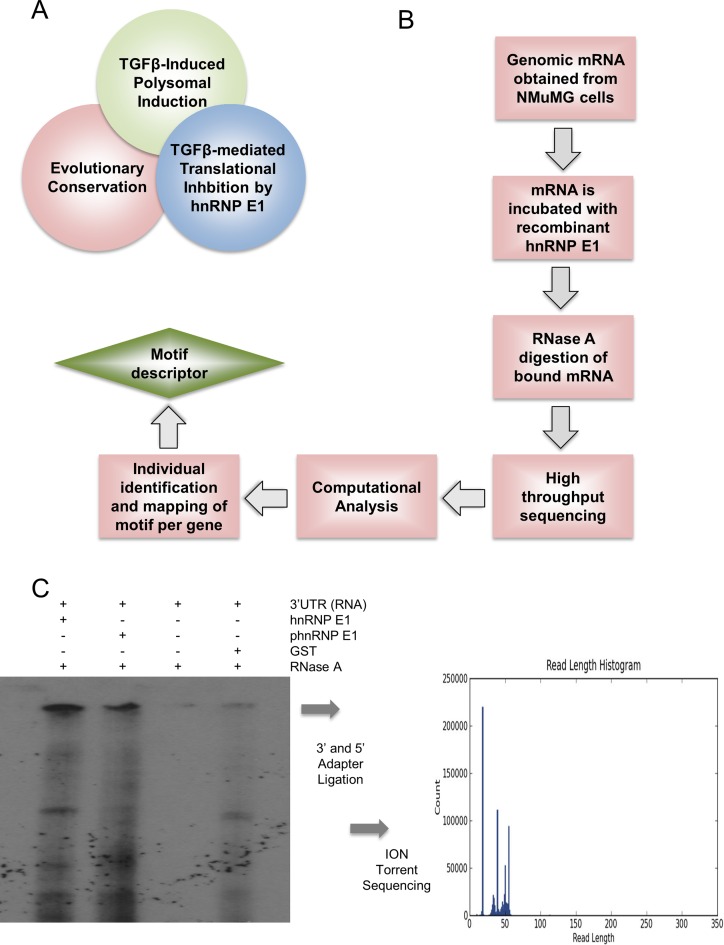
Figure 4.
Unbiased exonuclease digestion of hnRNP E1 bound target mRNA allows for high-throughput analysis of protected RNA fragment sequences. (A) Venn diagram depiction of genomic characteristics displayed by p-hnRNP E1 responsive genes. (B) Flow diagram of unbiased, genomic approach for resolving 3′-UTR consensus motif. This modification to common exonuclease assays utilizes homologous recombinant proteins for the sake of lowered background and increased specificity of sequencing results. (C) Exonuclease digestion assay for genomic RNA obtained from NMuMG cells (using trizol extraction) and bound to either hnRNP E1 or p-hnRNP E1. p-hnRNP E1 serves as a control to account for the promiscuous nature of hnRNP E1 for binding other RNAs that are not specific to its serine-43 reversible binding region. A unique band found only in samples protected by hnRNP E1 was ligated with 3′ and 5′ sequencing primers and sequenced using ION-torrent sequencing.