Figure 1.
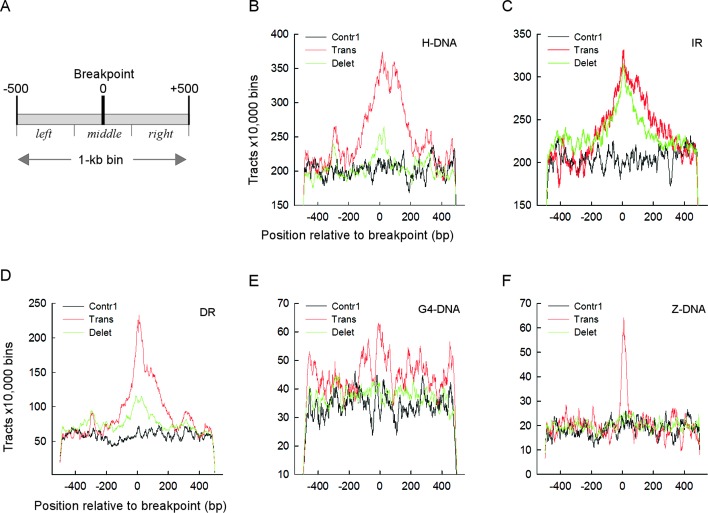
Translocation and deletion breakpoints occur near PONDS-forming motifs. (A) Schematic of a 1 kb-bin showing the breakpoint at position 0 and three sections: left from −500 to −177; middle from −176 to 176; and right from 177 to 500. (B) Number of DNA triplex-forming repeats (H-DNA) for 10 000 bins found near translocation (red), deletion (green) and Contr1 (black) breakpoints. (C) Same as in B, but for cruciform-forming inverted repeats (IR). (D) Same as in B, but for loop DNA-forming tandem repeats (DR). (E) Same as in B, but for quadruplex-forming repeats (G4-DNA). (F) Same as in B, but for left-handed DNA-forming repeats (Z-DNA). Numbers refers to the counts of bases belonging to each repeat type at every position; for H-DNA and IR, any bases separating a pair of repeats were excluded from the count.