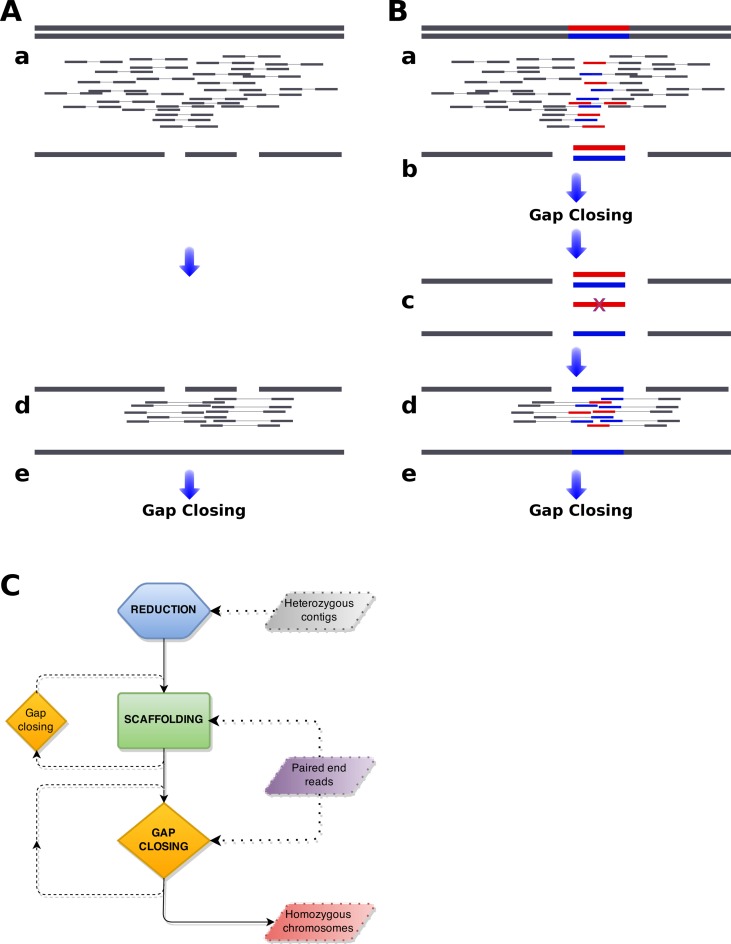
Figure 1.
Genome assembly from short reads. Standard (A) and heterozygous (B) genome assembly pipelines are compared. Diploid chromosomes are indicated as horizonal bars with heterozygous regions marked as red and blue. Paired-end reads produced from sequencing of those chromosomes are indicated as smaller bars linked by thin lines below the chromosomes. Assemblies are indicated as horizontal bars, in the same way as chromosomes, but a single reference is produced for diploid chromosomes. Heterozygous genome assembly pipeline consists of five steps. (a) Standard de novo assembly is performed and (b) optionally gaps are closed. Obtained assembly is larger than expected and fragmented because two alternative contigs are recovered from heterozygous regions (blue and red), while single contig is recovered from homozygous regions (gray). Further scaffolding of such assembly is impossible, as homozygous contigs can be joined to any of heterozygous contigs (blue and red).(c) To overcome this, redundant contigs from heterozygous regions are removed (here the red contig) and (d) homogenised assembly is further scaffolded. (e) Finally, gaps are closed.(C) Schematic representation of redundans mechanisms. Redundans pipeline consists of three steps: reduction, scaffolding and gap closing. Program takes as input assembled contigs, paired-end and/or mate pairs sequencing libraries and returns scaffolded homozygous genome assembly, that should be less fragmented and with total size smaller than the input contigs. Note, scaffolding and gap closing may be executed in multiple iterations. In the first step, only heterozygous contigs are used. Paired-end and/or mate pair libraries are used for scaffolding and gap closing. The latter steps can be repeated to achieve incremental assembly improvement. Redundans is very flexible, thus any of the above mentioned step can be omitted.