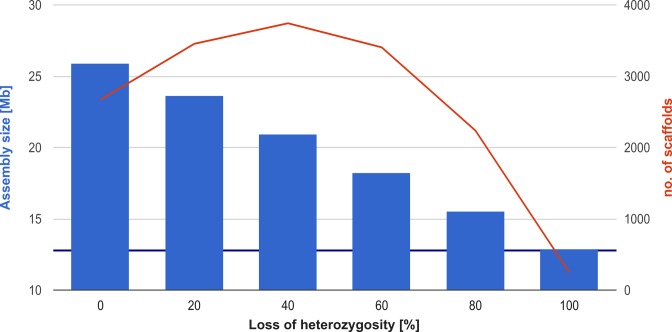
Figure 2.
Heterozygous genome assemblies characteristics. The total SOAPdenovo2 assembly size (blue bars) as well as number of scaffolds longer than 1 kb (red plot) are given for reconstructed genome assemblies: one homozygous (LOH of 100%) and five heterozygous (with 5% divergence between haplomes and varying loss of heterozygosity level: 0%, 20%, 40%, 60%, 80%). Expected genome size is marked with purple baseline. The assemblies recovered for heterozygous genomes are much more fragmented (2237–3743 scaffolds) than those recovered for homozygous genome (250 scaffolds). The assembly reconstructed for homozygous genome (100% LOH) has expected size, while the remaining assemblies have size larger than expected (119.20–198.85% of expected size). The size of the genome assembly is negatively correlated with LOH level (Pearson coefficient of -0.9996).