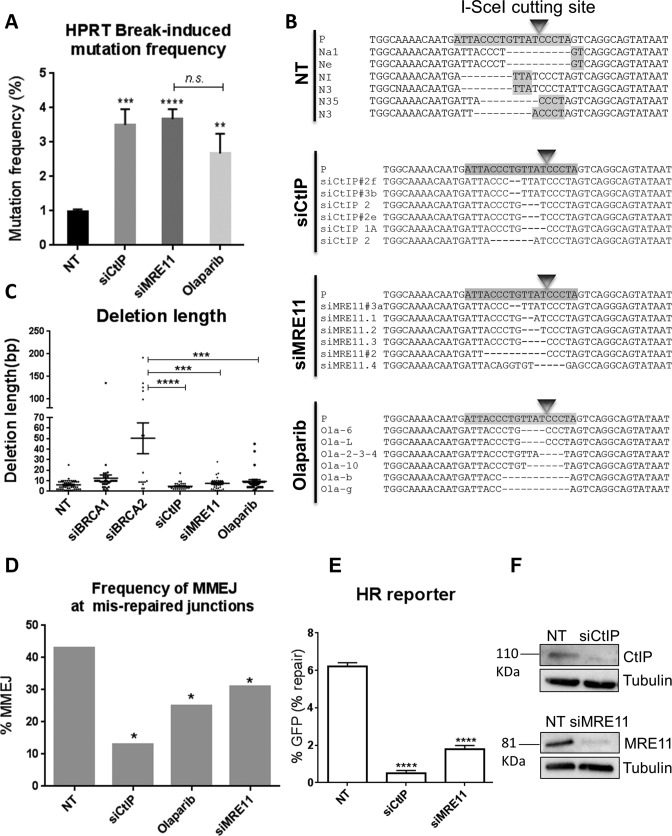
Figure 4.
DSB-induced mutation signatures in HR-deficient cells require MMEJ factors. (A) Break-induced mutation frequency of HPRT reporter cells treated with NT control siRNA (NT), CtIP siRNA (siCtIP), MRE11 siRNA (siMRE11) or PARP-1 inhibitor (Olaparib). Error bars represent SEM from three independent experiments, n.s., not significant, **P < 0.01, ***P < 0.001, ****P < 0.0001. (B) Representative sequence alignments of the HPRT negative PCR products in cells treated with NT control siRNA (NT), CtIP siRNA (siCtIP), MRE11 siRNA (siMRE11) or PARP-1 inhibitor (Olaparib), from three independent experiments (see also Supplementary Figures S1A, S8–10). I-SceI recognition sequence and terminal microhomologies at the break sites are highlighted. (C) Average deletion lengths (bp) in different genetic backgrounds. Each dot represents an independent clone. The lines represent mean and SEM, ***P < 0.001, ****P < 0.0001. (D) Frequency of MMEJ at mis-repaired junctions in HPRT deletion mutants isolated from cells treated with NT control siRNA (NT) or CtIP siRNA (siCtIP), MRE11 siRNA (siMRE11) or Olaparib. P values calculated by statistical analysis ‘‘difference between proportions’’, *P < 0.05. (E) HR repair efficacy of DR-GFP reporter cells treated with NT control siRNA (NT), CtIP siRNA (siCtIP), or MRE11 siRNA (siMRE11), indicated by the percentage of GFP-positive cells. Error bars show SEM from three independent experiments. ****P < 0.0001. (F) Western blot showing CtIP and MRE11 knockdowns 48 h following siRNA transfection.