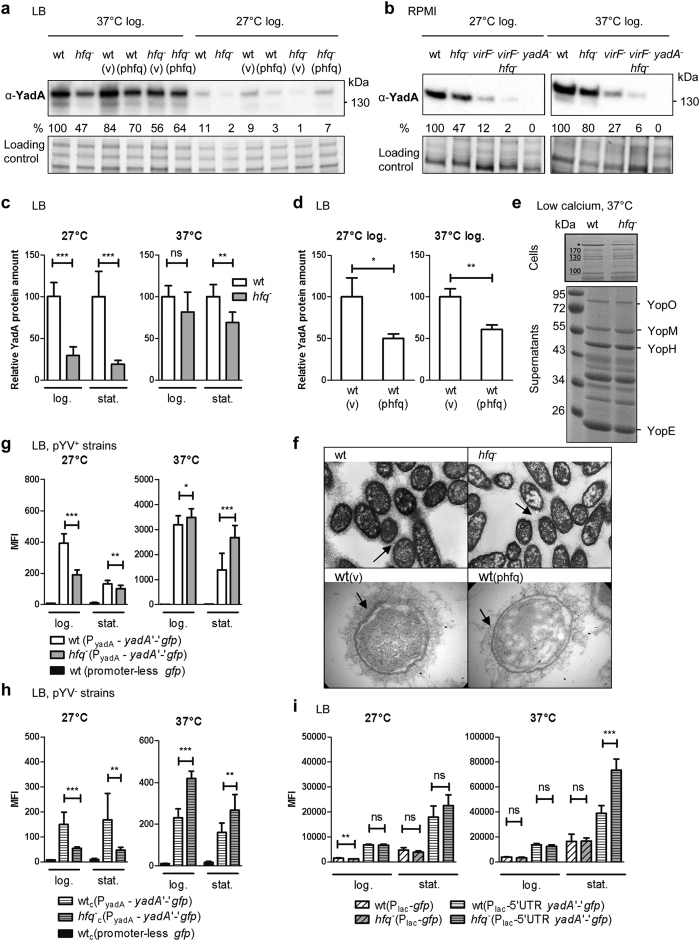
Figure 3. Complex regulation of adhesin YadA by Hfq.
(a–f) Absence of hfq, but also multiple copies of hfq, lead to decreased YadA abundance. (a) Immunodetection of YadA in Y. enterocolitica strain WA-314 and derivatives grown in LB for 4 h. Upper panel shows the immunoblot and bottom panel shows part of the gel stained for total proteins. Percentages indicate the relative YadA signal intensities. (b) Hfq promotes production of YadA, independently of the VirF transcriptional activator. Strain WA-314 and derivatives were grown in RPMI for 4 h. (c,d) Semi-quantitative analysis of total YadA protein amounts in parent and hfq mutant grown in LB for 4h (log.) or 20 h (stat.). Results are the relative mean signal and standard deviation obtained from six (c) or three (d) independent cultures loaded on the same gel. (e) Hfq is dispensable for T3SS but not YadA production. Coomassie blue-stained gels of cell extracts (top) and supernatants (bottom) prepared upon growth in T3SS-inducing conditions. The asterisk indicates the YadA trimer. (f) Hfq impacts deposition of the collagen fibrils associated to YadA at the bacterial surface. Electron microscope images of bacteria grown for 20 h at 37 °C in collagen gels. Original magnification: upper panels, ×15000; lower panels, ×21000. Arrows point to the YadA-dependent deposited collagen fibrils. (g and h) Hfq exerts opposite effects on yadA expression at 27 °C and 37 °C (g), also in the absence of the pYV plasmid (h). Fluorescence of strain WA-314 and derivatives carrying promoter-less gfp or the translational fusion yadA‘-’gfp under the control of promoter PyadA was measured by flow cytometry upon growth in LB for 4 h and 22 h. (i) Hfq inhibits yadA expression at the post-transcriptional level at 37 °C in stationary phase. Bacterial MFI of strains carrying pFX-1 (Plac–driven gfp) and pFX-Plac-yadA (yadA 5′UTR and yadA‘-’ gfp fusion under control of Plac). Results are the MFI and standard deviation of at least two independent experiments, each with three independent cultures per strain. Significance was calculated with Student’s unpaired t-test (***P ≤ 0.001; **P ≤ 0.01, *P ≤ 0.05; ns, not significant, P > 0.05).