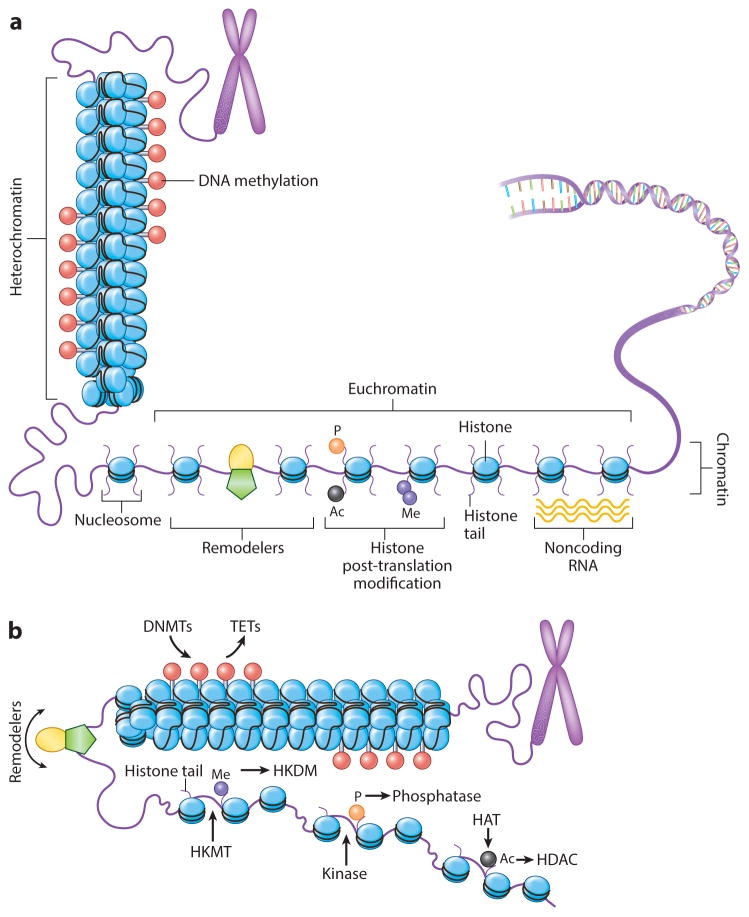
Figure 1.
The epigenome landscape. (a) Chromatin states support either transcriptional activation or silencing of genes, allowing gene regulatory regions to switch these states through positioning of nucleosomes (blue ovals). More open conformations leave the transcription start site nucleosome free. Modifications of nucleosome histone tails ( purple lines extending from ovals) regulate the process, including DNA methylation (red lollipops), serine phosphorylation (orange circle), lysine acetylation (black circle) and lysine methylation ( purple circle), and nucleosome remodeler complexes ( green pentagon with yellow oval). Additionally, noncoding RNAs ( yellow waves) can participate in these regulatory steps through recruitment of chromatin proteins and DNA methylation. (b) Control of histone modifications and of DNA methylation by proteins: writers (DNMTs, HKMTs, HATs, kinases for phosphorylation), readers (shown in subsequent figures for binding to and interpreting each mark for function), erasers (TETs for DNA methylation, HKDMs for lysine methylation, HDACs, phosphatases for removing phosphorylation) and nucleosome remodelers. Red lollipops indicate DNA methylation; green pentagon with yellow oval indicates nucleosome remodeler complexes; purple circle indicates histone lysine methylation; orange circle indicates serine phosphorylation; black circle indicates lysine acetylation. Abbreviations: DNMT, DNA methyltransferase; HAT, histone acetylases; HDAC, histone deacetylases; HKDM, histone lysine demethylase; HKMT, histone lysine methyltransferase; TET, ten-eleven translocation protein.